Targeting bromodomain and extraterminal proteins in breast cancer
- PMID: 29154989
- PMCID: PMC5828951
- DOI: 10.1016/j.phrs.2017.11.015
Targeting bromodomain and extraterminal proteins in breast cancer
Abstract
Breast cancer is a collection of distinct tumor subtypes that are driven by unique gene expression profiles. These transcriptomes are controlled by various epigenetic marks that dictate which genes are expressed and suppressed. During carcinogenesis, extensive restructuring of the epigenome occurs, including aberrant acetylation, alteration of methylation patterns, and accumulation of epigenetic readers at oncogenes. As epigenetic alterations are reversible, epigenome-modulating drugs could provide a mechanism to silence numerous oncogenes simultaneously. Here, we review the impact of inhibitors of the Bromodomain and Extraterminal (BET) family of epigenetic readers in breast cancer. These agents, including the prototypical BET inhibitor JQ1, have been shown to suppress a variety of oncogenic pathways while inducing minimal, if any, toxicity in models of several subtypes of breast cancer. BET inhibitors also synergize with multiple approved anti-cancer drugs, providing a greater response in breast cancer cell lines and mouse models than either single agent. The combined findings of the studies discussed here provide an excellent rationale for the continued investigation of the utility of BET inhibitors in breast cancer.
Keywords: ARV-825 PubChem CID 92044400; Anti-cancer drugs; BET inhibitor; BETd-246 PubChem CID 131698640; Breast cancer; Bromodomain and extraterminal protein; Drug synergy; I-BET151 PubChem CID 52912189; I-BET762 PubChem CID 46943432; JQ1 PubChem CID 46907787; MS417 PubChem CID 59190723; OTX015 PubChem CID 9936746; THZ1 PubChem CID 73602827; Transcriptional control; XD14 PubChem CID 52670832; dBET1 PubChem CID 91799313.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Figures
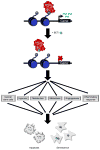
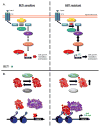

References
-
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge Ø, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
-
- Prat A, Pineda E, Adamo B, Galván P, Fernández A, Gaba L, Díez M, Viladot M, Arance A, Muñoz M. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015;24:S26–35. - PubMed
-
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE, Børresen-Dale A-L. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

