GPCRdb in 2018: adding GPCR structure models and ligands
- PMID: 29155946
- PMCID: PMC5753179
- DOI: 10.1093/nar/gkx1109
GPCRdb in 2018: adding GPCR structure models and ligands
Abstract
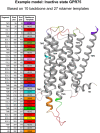
G protein-coupled receptors are the most abundant mediators of both human signalling processes and therapeutic effects. Herein, we report GPCRome-wide homology models of unprecedented quality, and roughly 150 000 GPCR ligands with data on biological activities and commercial availability. Based on the strategy of 'Less model - more Xtal', each model exploits both a main template and alternative local templates. This achieved higher similarity to new structures than any of the existing resources, and refined crystal structures with missing or distorted regions. Models are provided for inactive, intermediate and active states-except for classes C and F that so far only have inactive templates. The ligand database has separate browsers for: (i) target selection by receptor, family or class, (ii) ligand filtering based on cross-experiment activities (min, max and mean) or chemical properties, (iii) ligand source data and (iv) commercial availability. SMILES structures and activity spreadsheets can be downloaded for further processing. Furthermore, three recent landmark publications on GPCR drugs, G protein selectivity and genetic variants have been accompanied with resources that now let readers view and analyse the findings themselves in GPCRdb. Altogether, this update will enable scientific investigation for the wider GPCR community. GPCRdb is available at http://www.gpcrdb.org.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Fredriksson R., Lagerström M.C., Lundin L.-G., Schiöth H.B.. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003; 63:1256–1272. - PubMed
-
- Kolakowski L.F., Jr GCRDb: a G-protein-coupled receptor database. Receptors Channels. 1994; 2:1–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

