DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions
- PMID: 29156006
- PMCID: PMC5753203
- DOI: 10.1093/nar/gkx1141
DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions
Abstract
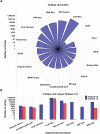
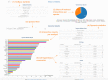
DIANA-TarBase v8 (http://www.microrna.gr/tarbase) is a reference database devoted to the indexing of experimentally supported microRNA (miRNA) targets. Its eighth version is the first database indexing >1 million entries, corresponding to ∼670 000 unique miRNA-target pairs. The interactions are supported by >33 experimental methodologies, applied to ∼600 cell types/tissues under ∼451 experimental conditions. It integrates information on cell-type specific miRNA-gene regulation, while hundreds of thousands of miRNA-binding locations are reported. TarBase is coming of age, with more than a decade of continuous support in the non-coding RNA field. A new module has been implemented that enables the browsing of interactions through different filtering combinations. It permits easy retrieval of positive and negative miRNA targets per species, methodology, cell type and tissue. An incorporated ranking system is utilized for the display of interactions based on the robustness of their supporting methodologies. Statistics, pie-charts and interactive bar-plots depicting the database content are available through a dedicated result page. An intuitive interface is introduced, providing a user-friendly application with flexible options to different queries.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Vlachos I.S., Hatzigeorgiou A.G.. Online resources for miRNA analysis. Clin. Biochem. 2013; 46:879–900. - PubMed
-
- Huntzinger E., Izaurralde E.. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011; 12:99–110. - PubMed
-
- Vlachos I.S., Georgakilas G., Tastsoglou S., Paraskevopoulou M.D., Karagkouni D., Hatzigeorgiou A.G.. De Pietri Tonelli D. Computational challenges and -omics approaches for the identification of miRNAs and targets. Essentials of Noncoding RNA in Neuroscience. 2017; Boston: Academic Press; 39–60.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

