Vemurafenib-resistance via de novo RBM genes mutations and chromosome 5 aberrations is overcome by combined therapy with palbociclib in thyroid carcinoma with BRAFV600E
- PMID: 29156680
- PMCID: PMC5689570
- DOI: 10.18632/oncotarget.21262
Vemurafenib-resistance via de novo RBM genes mutations and chromosome 5 aberrations is overcome by combined therapy with palbociclib in thyroid carcinoma with BRAFV600E
Abstract
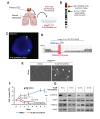
Purpose: Papillary thyroid carcinoma (PTC) is the most frequent endocrine tumor. BRAFV600E represents the PTC hallmark and is targeted with selective inhibitors (e.g. vemurafenib). Although there have been promising results in clinical trials using these inhibitors, most patients develop resistance and progress. Tumor clonal diversity is proposed as one mechanism underlying drug resistance. Here we have investigated mechanisms of primary and secondary resistance to vemurafenib in BRAFWT/V600E-positive PTC patient-derived cells with P16-/- (CDKN2A-/-).
Experimental design: Following treatment with vemurafenib, we expanded a sub-population of cells with primary resistance and characterized them genetically and cytogenetically. We have used exome sequencing, metaphase chromosome analysis, FISH and oligonucleotide SNP-microarray assays to assess clonal evolution of vemurafenib-resistant cells. Furthermore, we have validated our findings by networks and pathways analyses using PTC clinical samples.
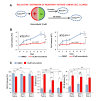
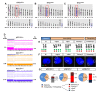
Results: Vemurafenib-resistant cells grow similarly to naïve cells but are refractory to apoptosis upon treatment with vemurafenib, and accumulate in G2-M phase. We find that vemurafenib-resistant cells show amplification of chromosome 5 and de novo mutations in the RBM (RNA-binding motifs) genes family (i.e. RBMX, RBM10). RBMX knockdown in naïve-cells contributes to tetraploidization, including expansion of clones with chromosome 5 aberrations (e.g. isochromosome 5p). RBMX elicits gene regulatory networks with chromosome 5q cancer-associated genes and pathways for G2-M and DNA damage-response checkpoint regulation in BRAFWT/V600E-PTC. Importantly, combined therapy with vemurafenib plus palbociclib (inhibitor of CDK4/6, mimicking P16 functions) synergistically induces stronger apoptosis than single agents in resistant-cells and in anaplastic thyroid tumor cells harboring the heterozygous BRAFWT/V600E mutation.
Conclusions: Critically, our findings suggest for the first time that targeting BRAFWT/V600E and CDK4/6 represents a novel therapeutic strategy to treat vemurafenib-resistant or vemurafenib-naïve radioiodine-refractory BRAFWT/V600E-PTC. This combined therapy could prevent selection and expansion of aggressive PTC cell sub-clones with intrinsic resistance, targeting tumor cells either with primary or secondary resistance to BRAFV600E inhibitor.
Keywords: BRAFV600E; chromosome 5; combined therapy with vemurafenib and palbociclib; drug resistance; papillary thyroid cancer preclinical model.
Conflict of interest statement
CONFLICTS OF INTEREST All authors declare no conflicts of interest that influence this manuscript.
Figures



Comment in
-
The role of palbociclib in thyroid carcinoma with BRAF mutation.Gland Surg. 2018 Aug;7(Suppl 1):S82-S85. doi: 10.21037/gs.2018.03.07. Gland Surg. 2018. PMID: 30175070 Free PMC article. No abstract available.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

