Independent impacts of aging on mitochondrial DNA quantity and quality in humans
- PMID: 29157198
- PMCID: PMC5697406
- DOI: 10.1186/s12864-017-4287-0
Independent impacts of aging on mitochondrial DNA quantity and quality in humans
Abstract
Background: The accumulation of mitochondrial DNA (mtDNA) mutations, and the reduction of mtDNA copy number, both disrupt mitochondrial energetics, and may contribute to aging and age-associated phenotypes. However, there are few genetic and epidemiological studies on the spectra of blood mtDNA heteroplasmies, and the distribution of mtDNA copy numbers in different age groups and their impact on age-related phenotypes. In this work, we used whole-genome sequencing data of isolated peripheral blood mononuclear cells (PBMCs) from the UK10K project to investigate in parallel mtDNA heteroplasmy and copy number in 1511 women, between 17 and 85 years old, recruited in the TwinsUK cohorts.
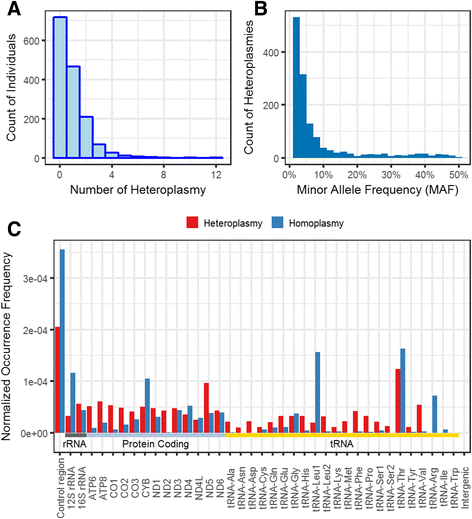
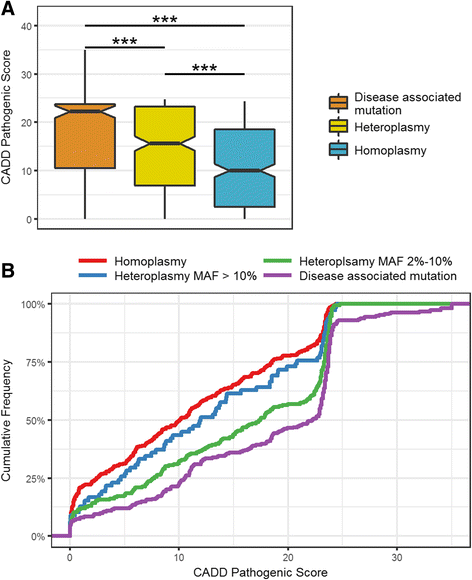
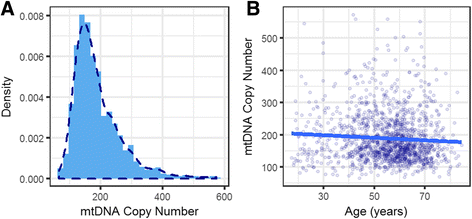
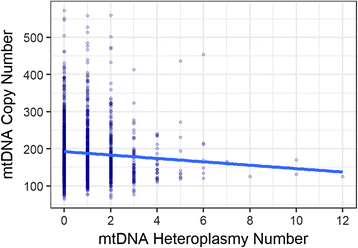
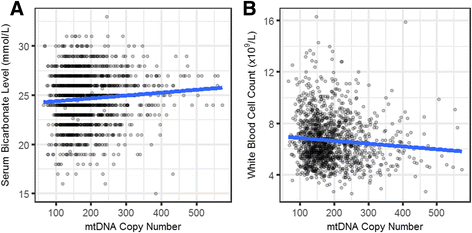
Results: We report a high prevalence of pathogenic mtDNA heteroplasmies in this population. We also find an increase in mtDNA heteroplasmies with age (β = 0.011, P = 5.77e-6), and showed that, on average, individuals aged 70-years or older had 58.5% more mtDNA heteroplasmies than those under 40-years old. Conversely, mtDNA copy number decreased by an average of 0.4 copies per year (β = -0.395, P = 0.0097). Multiple regression analyses also showed that age had independent effects on mtDNA copy number decrease and heteroplasmy accumulation. Finally, mtDNA copy number was positively associated with serum bicarbonate level (P = 4.46e-5), and inversely correlated with white blood cell count (P = 0.0006). Moreover, the aggregated heteroplasmy load was associated with blood apolipoprotein B level (P = 1.33e-5), linking the accumulation of mtDNA mutations to age-related physiological markers.
Conclusions: Our population-based study indicates that both mtDNA quality and quantity are influenced by age. An open question for the future is whether interventions that would contribute to maintain optimal mtDNA copy number and prevent the expansion of heteroplasmy could promote healthy aging.
Keywords: Aging; Heteroplasmy; Whole genome sequencing; mtDNA copy number.
Conflict of interest statement
Ethics approval and consent to participate
All subjects from TwinsUK cohort have ethical approval from the Guy’s and St Thomas’ (GSTT) Ethics Committee (Guy’s and St Thomas’ NHS Foundation Trust), and are compliant with the UK10K Ethical Governance Framework (
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

