Receptor-mediated Drp1 oligomerization on endoplasmic reticulum
- PMID: 29158231
- PMCID: PMC5716263
- DOI: 10.1083/jcb.201610057
Receptor-mediated Drp1 oligomerization on endoplasmic reticulum
Abstract
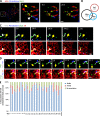
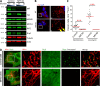
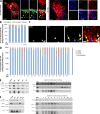
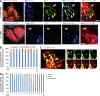
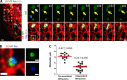
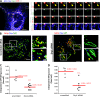
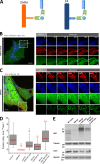
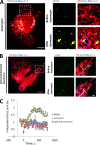
Drp1 is a dynamin guanosine triphosphatase important for mitochondrial and peroxisomal division. Drp1 oligomerization and mitochondrial recruitment are regulated by multiple factors, including interaction with mitochondrial receptors such as Mff, MiD49, MiD51, and Fis. In addition, both endoplasmic reticulum (ER) and actin filaments play positive roles in mitochondrial division, but mechanisms for their roles are poorly defined. Here, we find that a population of Drp1 oligomers is associated with ER in mammalian cells and is distinct from mitochondrial or peroxisomal Drp1 populations. Subpopulations of Mff and Fis1, which are tail-anchored proteins, also localize to ER. Drp1 oligomers assemble on ER, from which they can transfer to mitochondria. Suppression of Mff or inhibition of actin polymerization through the formin INF2 significantly reduces all Drp1 oligomer populations (mitochondrial, peroxisomal, and ER bound) and mitochondrial division, whereas Mff targeting to ER has a stimulatory effect on division. Our results suggest that ER can function as a platform for Drp1 oligomerization, and that ER-associated Drp1 contributes to mitochondrial division.
© 2017 Ji et al.
Figures

References
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

