Deep sequencing of HIV-1 reverse transcripts reveals the multifaceted antiviral functions of APOBEC3G
- PMID: 29158605
- PMCID: PMC6014619
- DOI: 10.1038/s41564-017-0063-9
Deep sequencing of HIV-1 reverse transcripts reveals the multifaceted antiviral functions of APOBEC3G
Abstract
Following cell entry, the RNA genome of HIV-1 is reverse transcribed into double-stranded DNA that ultimately integrates into the host-cell genome to establish the provirus. These early phases of infection are notably vulnerable to suppression by a collection of cellular antiviral effectors, called restriction or resistance factors. The host antiviral protein APOBEC3G (A3G) antagonizes the early steps of HIV-1 infection through the combined effects of inhibiting viral cDNA production and cytidine-to-uridine-driven hypermutation of this cDNA. In seeking to address the underlying molecular mechanism for inhibited cDNA synthesis, we developed a deep sequencing strategy to characterize nascent reverse transcription products and their precise 3'-termini in HIV-1 infected T cells. Our results demonstrate site- and sequence-independent interference with reverse transcription, which requires the specific interaction of A3G with reverse transcriptase itself. This approach also established, contrary to current ideas, that cellular uracil base excision repair (UBER) enzymes target and cleave A3G-edited uridine-containing viral cDNA. Together, these findings yield further insights into the regulatory interplay between reverse transcriptase, A3G and cellular DNA repair machinery, and identify the suppression of HIV-1 reverse transcriptase by a directly interacting host protein as a new cell-mediated antiviral mechanism.
Conflict of interest statement
The authors have no conflict of financial interest.
Figures

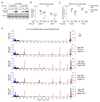
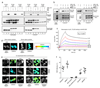



Comment in
-
A multimodal antiretroviral protein.Nat Microbiol. 2018 Feb;3(2):122-123. doi: 10.1038/s41564-017-0104-4. Nat Microbiol. 2018. PMID: 29358682 No abstract available.
References
-
- Bishop KN, et al. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

