Computational and theoretical approaches for studies of a lipid recognition protein on biological membranes
- PMID: 29159013
- PMCID: PMC5689545
- DOI: 10.2142/biophysico.14.0_153
Computational and theoretical approaches for studies of a lipid recognition protein on biological membranes
Abstract
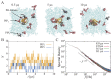
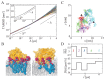
Many cellular functions, including cell signaling and related events, are regulated by the association of peripheral membrane proteins (PMPs) with biological membranes containing anionic lipids, e.g., phosphatidylinositol phosphate (PIP). This association is often mediated by lipid recognition modules present in many PMPs. Here, I summarize computational and theoretical approaches to investigate the molecular details of the interactions and dynamics of a lipid recognition module, the pleckstrin homology (PH) domain, on biological membranes. Multiscale molecular dynamics simulations using combinations of atomistic and coarse-grained models yielded results comparable to those of actual experiments and could be used to elucidate the molecular mechanisms of the formation of protein/lipid complexes on membrane surfaces, which are often difficult to obtain using experimental techniques. Simulations revealed some modes of membrane localization and interactions of PH domains with membranes in addition to the canonical binding mode. In the last part of this review, I address the dynamics of PH domains on the membrane surface. Local PIP clusters formed around the proteins exhibit anomalous fluctuations. This dynamic change in protein-lipid interactions cause temporally fluctuating diffusivity of proteins, i.e., the short-term diffusivity of the bound protein changes substantially with time, and may in turn contribute to the formation/dissolution of protein complexes in membranes.
Keywords: Peripheral membrane protein; molecular dynamics simulation; phosphatidylinositol phosphate; pleckstrin homology domain; protein-lipid interaction.
Conflict of interest statement
Conflict of Interest The author declares no conflicts of interest.
Figures

Similar articles
-
Dynamic interactions between a membrane binding protein and lipids induce fluctuating diffusivity.Sci Adv. 2017 Jan 20;3(1):e1601871. doi: 10.1126/sciadv.1601871. eCollection 2017 Jan. Sci Adv. 2017. PMID: 28116358 Free PMC article.
-
Anomalous Dynamics of a Lipid Recognition Protein on a Membrane Surface.Sci Rep. 2015 Dec 14;5:18245. doi: 10.1038/srep18245. Sci Rep. 2015. PMID: 26657413 Free PMC article.
-
Modes of Interaction of Pleckstrin Homology Domains with Membranes: Toward a Computational Biochemistry of Membrane Recognition.J Mol Biol. 2018 Feb 2;430(3):372-388. doi: 10.1016/j.jmb.2017.12.011. Epub 2017 Dec 20. J Mol Biol. 2018. PMID: 29273202
-
The energetics of protein-lipid interactions as viewed by molecular simulations.Biochem Soc Trans. 2020 Feb 28;48(1):25-37. doi: 10.1042/BST20190149. Biochem Soc Trans. 2020. PMID: 31872229 Free PMC article. Review.
-
Cellular and molecular interactions of phosphoinositides and peripheral proteins.Chem Phys Lipids. 2014 Sep;182:3-18. doi: 10.1016/j.chemphyslip.2014.02.002. Epub 2014 Feb 17. Chem Phys Lipids. 2014. PMID: 24556335 Free PMC article. Review.
Cited by
-
Regulation of actin assembly by PI(4,5)P2 and other inositol phospholipids: An update on possible mechanisms.Biochem Biophys Res Commun. 2018 Nov 25;506(2):307-314. doi: 10.1016/j.bbrc.2018.07.155. Epub 2018 Aug 13. Biochem Biophys Res Commun. 2018. PMID: 30139519 Free PMC article. Review.
-
Characterization and computational simulation of human Syx, a RhoGEF implicated in glioblastoma.FASEB J. 2022 Jul;36(7):e22378. doi: 10.1096/fj.202101808RR. FASEB J. 2022. PMID: 35639414 Free PMC article.
References
-
- Cho W, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct. 2005;34:119–151. - PubMed
-
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. - PubMed
-
- Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. - PubMed
-
- Edidin M. Shrinking patches and slippery rafts: scales of domains in the plasma membrane. Trends Cell Biol. 2001;11:492–496. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
