Nucleic acid purification from plants, animals and microbes in under 30 seconds
- PMID: 29161268
- PMCID: PMC5697807
- DOI: 10.1371/journal.pbio.2003916
Nucleic acid purification from plants, animals and microbes in under 30 seconds
Erratum in
-
Correction: Nucleic acid purification from plants, animals and microbes in under 30 seconds.PLoS Biol. 2018 May 7;16(5):e1002630. doi: 10.1371/journal.pbio.1002630. eCollection 2018 May. PLoS Biol. 2018. PMID: 29734341 Free PMC article.
Abstract
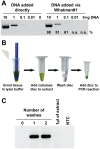
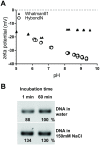
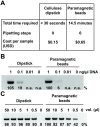
Nucleic acid amplification is a powerful molecular biology tool, although its use outside the modern laboratory environment is limited due to the relatively cumbersome methods required to extract nucleic acids from biological samples. To address this issue, we investigated a variety of materials for their suitability for nucleic acid capture and purification. We report here that untreated cellulose-based paper can rapidly capture nucleic acids within seconds and retain them during a single washing step, while contaminants present in complex biological samples are quickly removed. Building on this knowledge, we have successfully created an equipment-free nucleic acid extraction dipstick methodology that can obtain amplification-ready DNA and RNA from plants, animals, and microbes from difficult biological samples such as blood and leaves from adult trees in less than 30 seconds. The simplicity and speed of this method as well as the low cost and availability of suitable materials (e.g., common paper towelling), means that nucleic acid extraction is now more accessible and affordable for researchers and the broader community. Furthermore, when combined with recent advancements in isothermal amplification and naked eye DNA visualization techniques, the dipstick extraction technology makes performing molecular diagnostic assays achievable in limited resource settings including university and high school classrooms, field-based environments, and developing countries.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Ward E, Foster SJ, Fraaije BA, McCartney HA. Plant pathogen diagnostics: immunological and nucleic acid-based approaches. Ann Appl Biol. 2004;145: 1–16.
-
- Allen G, Flores-Vergara M, Krasynanski S, Kumar S, Thompson W. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protocols. 2006;1: 2320–2325. doi: 10.1038/nprot.2006.384 - DOI - PubMed
-
- Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. Vol. 3 Cold Springs Laboratory Press; Cold Spring Harbor, New York; 2001.
-
- Tan SC, Yiap BC. DNA, RNA, and protein extraction: the past and the present. BioMed Research International. 2009;2009: Article ID 574398. doi: 10.1155/2009/574398 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

