Fasciola spp: Mapping of the MF6 epitope and antigenic analysis of the MF6p/HDM family of heme-binding proteins
- PMID: 29161330
- PMCID: PMC5697881
- DOI: 10.1371/journal.pone.0188520
Fasciola spp: Mapping of the MF6 epitope and antigenic analysis of the MF6p/HDM family of heme-binding proteins
Abstract
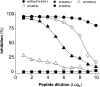
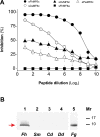
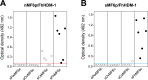
MF6p/FhHDM-1 is a small cationic heme-binding protein which is recognized by the monoclonal antibody (mAb) MF6, and abundantly present in parenchymal cells and secreted antigens of Fasciola hepatica. Orthologs of this protein (MF6p/HDMs) also exist in other causal agents of important foodborne trematodiasis, such as Clonorchis sinensis, Opisthorchis viverrini and Paragonimus westermani. Considering that MF6p/FhHDM-1 is relevant for heme homeostasis in Fasciola and was reported to have immunomodulatory properties, this protein is expected to be a useful target for vaccination. Thus, in this study we mapped the epitope recognized by mAb MF6 and evaluated its antigenicity in sheep. The sequence of the MF6p/FhHDM-1 ortholog from F. gigantica (MF6p/FgHDM-1) was also reported. By means of ELISA inhibitions with overlapping synthetic peptides, we determined that the epitope recognized by mAb MF6 is located within the C-terminal moiety of MF6p/FhHDM-1, which is the most conserved region of MF6p/HDMs. By immunoblotting analysis of parasite extracts and ELISA inhibitions with synthetic peptides we also determined that mAb MF6 reacted with the same intensity with F. hepatica and F. gigantica, and in decreasing order of intensity with C. sinensis, O.viverrini and P. westermani orthologs. On the contrary, mAb MF6 showed no reactivity against Dicrocoelium dendriticum and Schistosoma mansoni. The study of the recognition of peptides covering different regions of MF6p/FhHDM-1 by sera from immunized sheep revealed that the C-terminal moiety is the most antigenic, thus being of potential interest for vaccination. We also demonstrated that the production of antibodies to MF6p/FhHDM-1 in sheep infected by F. hepatica occurs relatively early and follows the same pattern as those produced against L-cathepsins.
Conflict of interest statement
Figures





Similar articles
-
The MF6p/FhHDM-1 major antigen secreted by the trematode parasite Fasciola hepatica is a heme-binding protein.J Biol Chem. 2014 Jan 17;289(3):1441-56. doi: 10.1074/jbc.M113.499517. Epub 2013 Nov 26. J Biol Chem. 2014. PMID: 24280214 Free PMC article.
-
Delineating distinct heme-scavenging and -binding functions of domains in MF6p/helminth defense molecule (HDM) proteins from parasitic flatworms.J Biol Chem. 2017 May 26;292(21):8667-8682. doi: 10.1074/jbc.M116.771675. Epub 2017 Mar 27. J Biol Chem. 2017. PMID: 28348084 Free PMC article.
-
Vaccination of sheep with Quil-A® adjuvant expands the antibody repertoire to the Fasciola MF6p/FhHDM-1 antigen and administered together impair the growth and antigen release of flukes.Vaccine. 2018 Apr 5;36(15):1949-1957. doi: 10.1016/j.vaccine.2018.02.115. Epub 2018 Mar 8. Vaccine. 2018. PMID: 29525280
-
Fasciola hepatica-Derived Molecules as Regulators of the Host Immune Response.Front Immunol. 2020 Sep 2;11:2182. doi: 10.3389/fimmu.2020.02182. eCollection 2020. Front Immunol. 2020. PMID: 32983184 Free PMC article. Review.
-
Immunomodulatory molecules of Fasciola hepatica: candidates for both vaccine and immunotherapeutic development.Vet Parasitol. 2013 Aug 1;195(3-4):272-85. doi: 10.1016/j.vetpar.2013.04.008. Epub 2013 Apr 6. Vet Parasitol. 2013. PMID: 23623183 Review.
Cited by
-
Pathogenicity and virulence of the liver flukes Fasciola hepatica and FasciolaGigantica that cause the zoonosis Fasciolosis.Virulence. 2021 Dec;12(1):2839-2867. doi: 10.1080/21505594.2021.1996520. Virulence. 2021. PMID: 34696693 Free PMC article.
-
Clonorchis sinensis MF6p/HDM (CsMF6p/HDM) induces pro-inflammatory immune response in RAW 264.7 macrophage cells via NF-κB-dependent MAPK pathways.Parasit Vectors. 2020 Jan 13;13(1):20. doi: 10.1186/s13071-020-3882-0. Parasit Vectors. 2020. PMID: 31931867 Free PMC article.
-
Increased specificity of Fasciola hepatica excretory-secretory antigens combining negative selection on hydroxyapatite and salt precipitation.Sci Rep. 2024 Feb 16;14(1):3897. doi: 10.1038/s41598-024-54290-8. Sci Rep. 2024. PMID: 38365880 Free PMC article.
-
Identification of protective peptides of Fasciola hepatica-derived cathepsin L1 (FhCL1) in vaccinated sheep by a linear B-cell epitope mapping approach.Parasit Vectors. 2020 Jul 31;13(1):390. doi: 10.1186/s13071-020-04260-6. Parasit Vectors. 2020. PMID: 32736582 Free PMC article.
-
Efficacy of a multivalent vaccine against Fasciola hepatica infection in sheep.Vet Res. 2021 Jan 28;52(1):13. doi: 10.1186/s13567-021-00895-0. Vet Res. 2021. PMID: 33509286 Free PMC article.
References
-
- Keiser J, Utzinger J. Emerging foodborne trematodiasis. Emerg Infect Dis. 2005;11:1507–1514. doi: 10.3201/eid1110.050614 - DOI - PMC - PubMed
-
- Mas-Coma S, Bargues MD, Valero MA. Fascioliasis and other plant-borne trematode zoonoses. Int J Parasitol. 2005;35:1255–1278. doi: 10.1016/j.ijpara.2005.07.010 - DOI - PubMed
-
- Mas-Coma S, Valero MA, Bargues MD. Fascioliasis. Adv Exp Med Biol. 2014;766:77–114. doi: 10.1007/978-1-4939-0915-5_4 - DOI - PubMed
-
- Valero MA, Bargues MD, Khoubbane M, Artigas P, Quesada C, Berinde L, et al. Higher physiopathogenicity by Fasciola gigantica than by the genetically close F. hepatica: experimental long-term follow-up of biochemical markers. Trans R Soc Trop Med Hyg. 2016;110:55–66. doi: 10.1093/trstmh/trv110 - DOI - PubMed
-
- Charlier J, Duchateau L, Claerebout E, Williams D, Vercruysse J. Associations between anti-Fasciola hepatica antibody levels in bulk-tank milk samples and production parameters in dairy herds. Prev Vet Med. 2007;78:57–66. doi: 10.1016/j.prevetmed.2006.09.010 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

