DNA polymerase beta participates in DNA End-joining
- PMID: 29161447
- PMCID: PMC5758893
- DOI: 10.1093/nar/gkx1147
DNA polymerase beta participates in DNA End-joining
Abstract
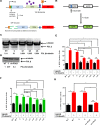
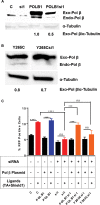
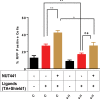
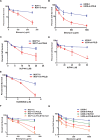
DNA double strand breaks (DSBs) are one of the most deleterious lesions and if left unrepaired, they lead to cell death, genomic instability and carcinogenesis. Cells combat DSBs by two pathways: homologous recombination (HR) and non-homologous end-joining (NHEJ), wherein the two DNA ends are re-joined. Recently a back-up NHEJ pathway has been reported and is referred to as alternative NHEJ (aNHEJ), which joins ends but results in deletions and insertions. NHEJ requires processing enzymes including nucleases and polymerases, although the roles of these enzymes are poorly understood. Emerging evidence indicates that X family DNA polymerases lambda (Pol λ) and mu (Pol μ) promote DNA end-joining. Here, we show that DNA polymerase beta (Pol β), another member of the X family of DNA polymerases, plays a role in aNHEJ. In the absence of DNA Pol β, fewer small deletions are observed. In addition, depletion of Pol β results in cellular sensitivity to bleomycin and DNA protein kinase catalytic subunit inhibitors due to defective repair of DSBs. In summary, our results indicate that Pol β in functions in aNHEJ and provide mechanistic insight into its role in this process.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

