To what extent do structural changes in catalytic metal sites affect enzyme function?
- PMID: 29161638
- PMCID: PMC5760197
- DOI: 10.1016/j.jinorgbio.2017.11.002
To what extent do structural changes in catalytic metal sites affect enzyme function?
Abstract
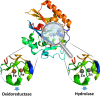
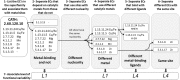
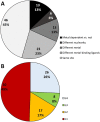
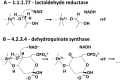
About half of known enzymatic reactions involve metals. Enzymes belonging to the same superfamily often evolve to catalyze different reactions on the same structural scaffold. The work presented here investigates how functional differentiation, within superfamilies that contain metalloenzymes, relates to structural changes at the catalytic metal site. In general, when the catalytic metal site is unchanged across the enzymes of a superfamily, the functional differentiation within the superfamily tends to be low and the mechanism conserved. Conversely, all types of structural changes in the metal binding site are observed for superfamilies with high functional differentiation. Overall, the catalytic role of the metal ions appears to be one of the most conserved features of the enzyme mechanism within metalloenzyme superfamilies. In particular, when the catalytic role of the metal ion does not involve a redox reaction (i.e. there is no exchange of electrons with the substrate), this role is almost always maintained even when the site undergoes significant structural changes. In these enzymes, functional diversification is most often associated with modifications in the surrounding protein matrix, which has changed so much that the enzyme chemistry is significantly altered. On the other hand, in more than 50% of the examples where the metal has a redox role in catalysis, changes at the metal site modify its catalytic role. Further, we find that there are no examples in our dataset where metal sites with a redox role are lost during evolution.
Synopsis: In this paper we investigate how functional diversity within superfamilies of metalloenzymes relates to structural changes at the catalytic metal site. Evolution tends to strictly conserve the metal site. When changes occur, they do not modify the catalytic role of non-redox metals whereas they affect the role of redox-active metals.
Keywords: Bioinorganic chemistry; Copper; Enzymes; Evolution; Iron; Magnesium; Metallo-proteins; Zinc.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Dayhoff M.O., Barker W.C., Hunt L.T. Methods Enzymol. 1983;91:524–545. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

