Production of highly efficient cellulase mixtures by genetically exploiting the potentials of Trichoderma reesei endogenous cellulases for hydrolysis of corncob residues
- PMID: 29162107
- PMCID: PMC5696804
- DOI: 10.1186/s12934-017-0825-3
Production of highly efficient cellulase mixtures by genetically exploiting the potentials of Trichoderma reesei endogenous cellulases for hydrolysis of corncob residues
Abstract
Background: Trichoderma reesei is one of the most important fungi utilized for cellulase production. However, its cellulase system has been proven to be present in suboptimal ratio for deconstruction of lignocellulosic substrates. Although previous enzymatic optimization studies have acquired different types of in vitro synthetic mixtures for efficient lignocellulose hydrolysis, production of in vivo optimized cellulase mixtures by industrial strains remains one of the obstacles to reduce enzyme cost in the biofuels production from lignocellulosic biomass.
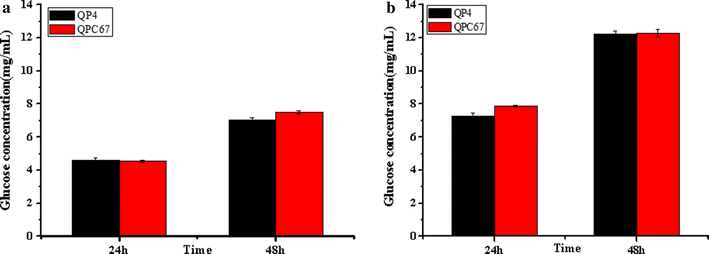
Results: In this study, we used a systematic genetic strategy based on the pyrG marker to overexpress the major cellulase components in a hypercellulolytic T. reesei strain and produce the highly efficient cellulase mixture for saccharification of corncob residues. We found that overexpression of CBH2 exhibited a 32-fold increase in the transcription level and a comparable protein level to CBH1, the most abundant secreted protein in T. reesei, but did not contribute much to the cellulolytic ability. However, when EG2 was overexpressed with a 46-fold increase in the transcription level and a comparable protein level to CBH2, the engineered strain QPE36 showed a 1.5-fold enhancement in the total cellulase activity (up to 5.8 U/mL FPA) and a significant promotion of saccharification efficiency towards differently pretreated corncob residues. To assist the following genetic manipulations, the marker pyrG was successfully excised by homologous recombination based on resistance to 5-FOA. Furthermore, BGL1 was overexpressed in the EG2 overexpression strain QE51 (pyrG-excised) and a 11.6-fold increase in BGL activity was obtained. The EG2-BGL1 double overexpression strain QEB4 displayed a remarkable enhancement of cellulolytic ability on pretreated corncob residues. Especially, a nearly complete cellulose conversion (94.2%) was found for the delignified corncob residues after 48 h enzymatic saccharification.
Conclusions: These results demonstrate that genetically exploiting the potentials of T. reesei endogenous cellulases to produce highly efficient cellulase mixtures is a powerful strategy to promote the saccharification efficiency, which will eventually facilitate cost reduction for lignocellulose-based biofuels.
Keywords: BGL1; CBH2; EG2; Genetic engineering; Optimization; Saccharification; Trichderma reesei.
Figures










Similar articles
-
Development of a powerful synthetic hybrid promoter to improve the cellulase system of Trichoderma reesei for efficient saccharification of corncob residues.Microb Cell Fact. 2022 Jan 4;21(1):5. doi: 10.1186/s12934-021-01727-8. Microb Cell Fact. 2022. PMID: 34983541 Free PMC article.
-
Production of the versatile cellulase for cellulose bioconversion and cellulase inducer synthesis by genetic improvement of Trichoderma reesei.Biotechnol Biofuels. 2017 Nov 15;10:272. doi: 10.1186/s13068-017-0963-1. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 29167702 Free PMC article.
-
Characterization and Strain Improvement of a Hypercellulytic Variant, Trichoderma reesei SN1, by Genetic Engineering for Optimized Cellulase Production in Biomass Conversion Improvement.Front Microbiol. 2016 Aug 29;7:1349. doi: 10.3389/fmicb.2016.01349. eCollection 2016. Front Microbiol. 2016. PMID: 27621727 Free PMC article.
-
Deciphering the molecular mechanisms behind cellulase production in Trichoderma reesei, the hyper-cellulolytic filamentous fungus.Biosci Biotechnol Biochem. 2016 Sep;80(9):1712-29. doi: 10.1080/09168451.2016.1171701. Epub 2016 Apr 14. Biosci Biotechnol Biochem. 2016. PMID: 27075508 Review.
-
Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives.J Ind Microbiol Biotechnol. 2008 May;35(5):377-391. doi: 10.1007/s10295-008-0327-8. Epub 2008 Mar 13. J Ind Microbiol Biotechnol. 2008. PMID: 18338189 Review.
Cited by
-
Production of cellulosic ethanol and value-added products from corn fiber.Bioresour Bioprocess. 2022 Aug 13;9(1):81. doi: 10.1186/s40643-022-00573-9. Bioresour Bioprocess. 2022. PMID: 38647596 Free PMC article. Review.
-
Synergistic Action of a Lytic Polysaccharide Monooxygenase and a Cellobiohydrolase from Penicillium funiculosum in Cellulose Saccharification under High-Level Substrate Loading.Appl Environ Microbiol. 2020 Nov 10;86(23):e01769-20. doi: 10.1128/AEM.01769-20. Print 2020 Nov 10. Appl Environ Microbiol. 2020. PMID: 32978122 Free PMC article.
-
Development of a powerful synthetic hybrid promoter to improve the cellulase system of Trichoderma reesei for efficient saccharification of corncob residues.Microb Cell Fact. 2022 Jan 4;21(1):5. doi: 10.1186/s12934-021-01727-8. Microb Cell Fact. 2022. PMID: 34983541 Free PMC article.
-
Tailoring the expression of Xyr1 leads to efficient production of lignocellulolytic enzymes in Trichoderma reesei for improved saccharification of corncob residues.Biotechnol Biofuels Bioprod. 2022 Dec 17;15(1):142. doi: 10.1186/s13068-022-02240-9. Biotechnol Biofuels Bioprod. 2022. PMID: 36528622 Free PMC article.
-
The ERAD Pathway Participates in Fungal Growth and Cellulase Secretion in Trichoderma reesei.J Fungi (Basel). 2023 Jan 4;9(1):74. doi: 10.3390/jof9010074. J Fungi (Basel). 2023. PMID: 36675895 Free PMC article.
References
-
- Somerville C. Next generation biofuels. AIP Conf Proc. 2015;1642:44–50. doi: 10.1063/1.4916167. - DOI
-
- Limayem A, Ricke SC. Lignocellulosic biomass for bioethanol production: current perspectives, potential issues and future prospects. Prog Energy Combust Sci. 2012;38(4):449–467. doi: 10.1016/j.pecs.2012.03.002. - DOI
-
- Jørgensen H, Kristensen JB, Felby C. Enzymatic conversion of lignocellulose into fermentable sugars: challenges and opportunities. Biofpr. 2007;1(2):119–134.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

