Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer
- PMID: 29162158
- PMCID: PMC5699172
- DOI: 10.1186/s12943-017-0743-3
Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer
Abstract
Background: Chemoresistance has long been recognized as a major obstacle in cancer therapy. Clarifying the underlying mechanism of chemoresistance would result in novel strategies to improve patient's response to chemotherapeutics.
Methods: lncRNA expression levels in gastric cancer (GC) cells was detected by quantitative real-time PCR (qPCR). MALAT1 shRNAs and overexpression vector were transfected into GC cells to down-regulate or up-regulate MALAT1 expression. In vitro and in vivo assays were performed to investigate the functional role of MALAT1 in autophagy associated chemoresistance.
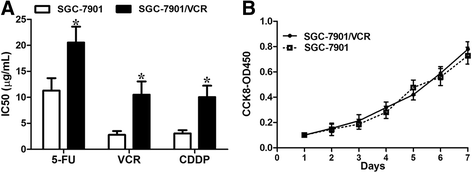
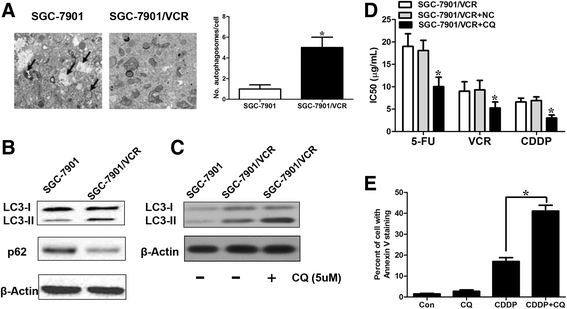
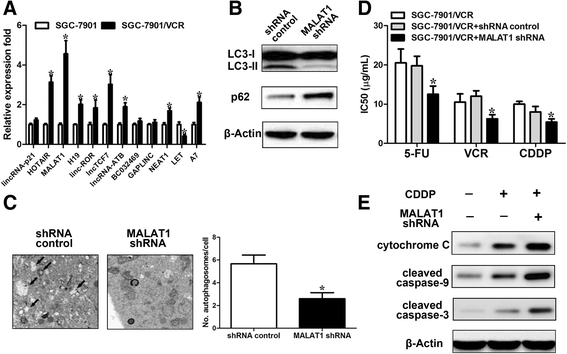
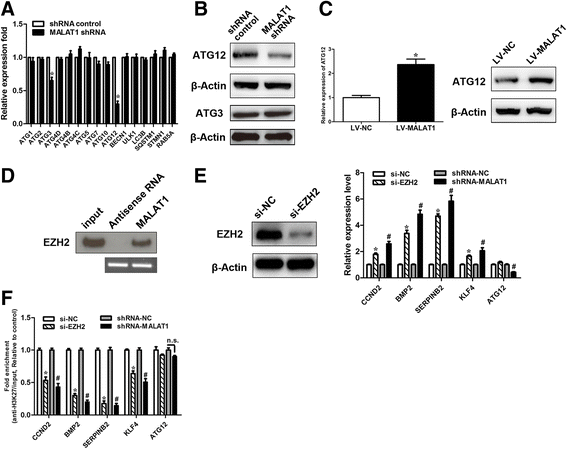
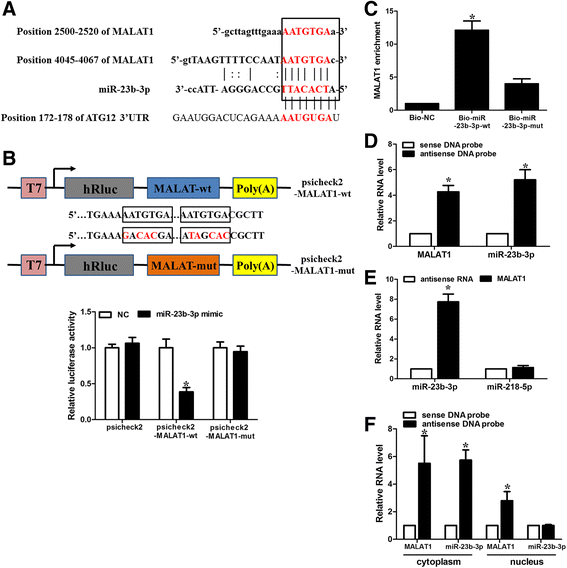
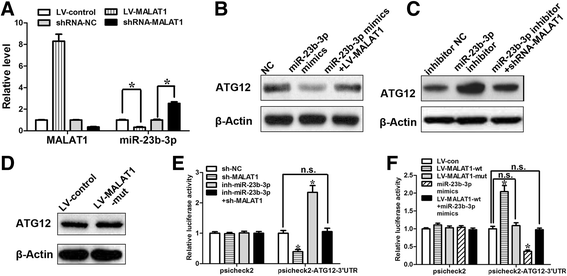
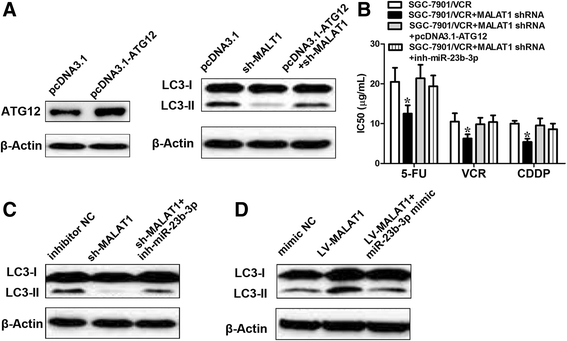
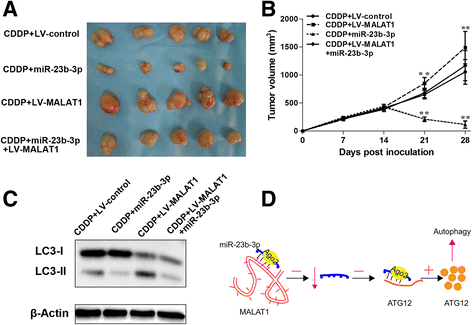
Results: We showed that chemoresistant GC cells had higher levels of MALAT1 and increased autophagy compared with parental cells. Silencing of MALAT1 inhibited chemo-induced autophagy, whereas MALAT1 promoted autophagy in gastric cancer cells. Knockdown of MALAT1 sensitized GC cells to chemotherapeutics. MALAT1 acts as a competing endogenous RNA for miR-23b-3p and attenuates the inhibitory effect of miR-23b-3p on ATG12, leading to chemo-induced autophagy and chemoresistance in GC cells.
Conclusions: Taken together, our study revealed a novel mechanism of lncRNA-regulated autophagy-related chemoresistance in GC, casting new lights on the understanding of chemoresistance.
Keywords: Autophagy; Chemoresistance; Gastric cancer; MALAT1; lncRNA.
Conflict of interest statement
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

