Mouse Model of Alagille Syndrome and Mechanisms of Jagged1 Missense Mutations
- PMID: 29162437
- PMCID: PMC7007299
- DOI: 10.1053/j.gastro.2017.11.002
Mouse Model of Alagille Syndrome and Mechanisms of Jagged1 Missense Mutations
Abstract
Background & aims: Alagille syndrome is a genetic disorder characterized by cholestasis, ocular abnormalities, characteristic facial features, heart defects, and vertebral malformations. Most cases are associated with mutations in JAGGED1 (JAG1), which encodes a Notch ligand, although it is not clear how these contribute to disease development. We aimed to develop a mouse model of Alagille syndrome to elucidate these mechanisms.
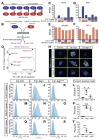
Methods: Mice with a missense mutation (H268Q) in Jag1 (Jag1+/Ndr mice) were outbred to a C3H/C57bl6 background to generate a mouse model for Alagille syndrome (Jag1Ndr/Ndr mice). Liver tissues were collected at different timepoints during development, analyzed by histology, and liver organoids were cultured and analyzed. We performed transcriptome analysis of Jag1Ndr/Ndr livers and livers from patients with Alagille syndrome, cross-referenced to the Human Protein Atlas, to identify commonly dysregulated pathways and biliary markers. We used species-specific transcriptome separation and ligand-receptor interaction assays to measure Notch signaling and the ability of JAG1Ndr to bind or activate Notch receptors. We studied signaling of JAG1 and JAG1Ndr via NOTCH 1, NOTCH2, and NOTCH3 and resulting gene expression patterns in parental and NOTCH1-expressing C2C12 cell lines.
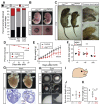
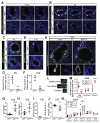
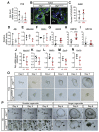
Results: Jag1Ndr/Ndr mice had many features of Alagille syndrome, including eye, heart, and liver defects. Bile duct differentiation, morphogenesis, and function were dysregulated in newborn Jag1Ndr/Ndr mice, with aberrations in cholangiocyte polarity, but these defects improved in adult mice. Jag1Ndr/Ndr liver organoids collapsed in culture, indicating structural instability. Whole-transcriptome sequence analyses of liver tissues from mice and patients with Alagille syndrome identified dysregulated genes encoding proteins enriched at the apical side of cholangiocytes, including CFTR and SLC5A1, as well as reduced expression of IGF1. Exposure of Notch-expressing cells to JAG1Ndr, compared with JAG1, led to hypomorphic Notch signaling, based on transcriptome analysis. JAG1-expressing cells, but not JAG1Ndr-expressing cells, bound soluble Notch1 extracellular domain, quantified by flow cytometry. However, JAG1 and JAG1Ndr cells each bound NOTCH2, and signaling from NOTCH2 signaling was reduced but not completely inhibited, in response to JAG1Ndr compared with JAG1.
Conclusions: In mice, expression of a missense mutant of Jag1 (Jag1Ndr) disrupts bile duct development and recapitulates Alagille syndrome phenotypes in heart, eye, and craniofacial dysmorphology. JAG1Ndr does not bind NOTCH1, but binds NOTCH2, and elicits hypomorphic signaling. This mouse model can be used to study other features of Alagille syndrome and organ development.
Keywords: Alagille; Development; Heart; Jagged1; Kidney; Liver; Notch; Vertebrae.
Copyright © 2018. Published by Elsevier Inc.
Conflict of interest statement
The authors disclose no conflicts. A separate project in ERA lab is funded by ModeRNA.
Figures


Comment in
-
Experimental model: A new mouse model of Alagille syndrome.Nat Rev Gastroenterol Hepatol. 2018 Jan;15(1):4. doi: 10.1038/nrgastro.2017.180. Epub 2017 Dec 13. Nat Rev Gastroenterol Hepatol. 2018. PMID: 29235546 No abstract available.
-
A New Model of Alagille Syndrome With Broad Phenotypic Representation.Gastroenterology. 2018 Mar;154(4):803-806. doi: 10.1053/j.gastro.2018.02.007. Epub 2018 Feb 6. Gastroenterology. 2018. PMID: 29425927 No abstract available.
References
-
- Mašek J, Andersson ER. The developmental biology of genetic Notch disorders. Development. 2017;144:1743–1763. - PubMed
-
- Li L, Krantz ID, Deng Y, et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251. - PubMed
-
- Oda T, Elkahloun AG, Pike BL, et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235–242. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

