Bacteriophage Transcytosis Provides a Mechanism To Cross Epithelial Cell Layers
- PMID: 29162715
- PMCID: PMC5698557
- DOI: 10.1128/mBio.01874-17
Bacteriophage Transcytosis Provides a Mechanism To Cross Epithelial Cell Layers
Erratum in
-
Correction for Nguyen et al., "Bacteriophage Transcytosis Provides a Mechanism To Cross Epithelial Cell Layers".mBio. 2018 Jan 2;9(1):e02207-17. doi: 10.1128/mBio.02207-17. mBio. 2018. PMID: 29295916 Free PMC article. No abstract available.
Abstract
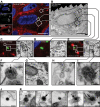
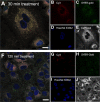
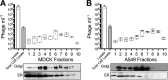
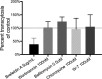
Bacterial viruses are among the most numerous biological entities within the human body. These viruses are found within regions of the body that have conventionally been considered sterile, including the blood, lymph, and organs. However, the primary mechanism that bacterial viruses use to bypass epithelial cell layers and access the body remains unknown. Here, we used in vitro studies to demonstrate the rapid and directional transcytosis of diverse bacteriophages across confluent cell layers originating from the gut, lung, liver, kidney, and brain. Bacteriophage transcytosis across cell layers had a significant preferential directionality for apical-to-basolateral transport, with approximately 0.1% of total bacteriophages applied being transcytosed over a 2-h period. Bacteriophages were capable of crossing the epithelial cell layer within 10 min with transport not significantly affected by the presence of bacterial endotoxins. Microscopy and cellular assays revealed that bacteriophages accessed both the vesicular and cytosolic compartments of the eukaryotic cell, with phage transcytosis suggested to traffic through the Golgi apparatus via the endomembrane system. Extrapolating from these results, we estimated that 31 billion bacteriophage particles are transcytosed across the epithelial cell layers of the gut into the average human body each day. The transcytosis of bacteriophages is a natural and ubiquitous process that provides a mechanistic explanation for the occurrence of phages within the body.IMPORTANCE Bacteriophages (phages) are viruses that infect bacteria. They cannot infect eukaryotic cells but can penetrate epithelial cell layers and spread throughout sterile regions of our bodies, including the blood, lymph, organs, and even the brain. Yet how phages cross these eukaryotic cell layers and gain access to the body remains unknown. In this work, epithelial cells were observed to take up and transport phages across the cell, releasing active phages on the opposite cell surface. Based on these results, we posit that the human body is continually absorbing phages from the gut and transporting them throughout the cell structure and subsequently the body. These results reveal that phages interact directly with the cells and organs of our bodies, likely contributing to human health and immunity.
Keywords: bacteriophages; endocytosis; phage-eukaryotic interaction; symbiosis; transcytosis.
Copyright © 2017 Nguyen et al.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
