Persistence and reversal of plasmid-mediated antibiotic resistance
- PMID: 29162798
- PMCID: PMC5698434
- DOI: 10.1038/s41467-017-01532-1
Persistence and reversal of plasmid-mediated antibiotic resistance
Abstract
In the absence of antibiotic-mediated selection, sensitive bacteria are expected to displace their resistant counterparts if resistance genes are costly. However, many resistance genes persist for long periods in the absence of antibiotics. Horizontal gene transfer (primarily conjugation) could explain this persistence, but it has been suggested that very high conjugation rates would be required. Here, we show that common conjugal plasmids, even when costly, are indeed transferred at sufficiently high rates to be maintained in the absence of antibiotics in Escherichia coli. The notion is applicable to nine plasmids from six major incompatibility groups and mixed populations carrying multiple plasmids. These results suggest that reducing antibiotic use alone is likely insufficient for reversing resistance. Therefore, combining conjugation inhibition and promoting plasmid loss would be an effective strategy to limit conjugation-assisted persistence of antibiotic resistance.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
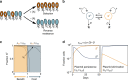
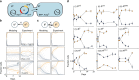

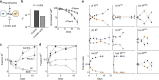
References
-
- Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 2010;8:260–271. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

