Intrinsic short-tailed azole resistance in mucormycetes is due to an evolutionary conserved aminoacid substitution of the lanosterol 14α-demethylase
- PMID: 29162893
- PMCID: PMC5698289
- DOI: 10.1038/s41598-017-16123-9
Intrinsic short-tailed azole resistance in mucormycetes is due to an evolutionary conserved aminoacid substitution of the lanosterol 14α-demethylase
Abstract
Mucormycoses are emerging and potentially lethal infections. An increase of breakthrough infections has been found in cohorts receiving short-tailed azoles prophylaxis (e.g. voriconazole (VCZ)). Although VCZ is ineffective in vitro and in vivo, long-tailed triazoles such as posaconazole remain active against mucormycetes. Our goal was to validate the molecular mechanism of resistance to short-tailed triazoles in Mucorales. The paralogous cytochrome P450 genes (CYP51 F1 and CYP51 F5) of Rhizopus arrhizus, Rhizopus microsporus, and Mucor circinelloides were amplified and sequenced. Alignment of the protein sequences of the R. arrhizus, R. microsporus, and M. circinelloides CYP51 F1 and F5 with additional Mucorales species (n = 3) and other fungi (n = 16) confirmed the sequences to be lanosterol 14α-demethylases (LDMs). Sequence alignment identified a pan-Mucorales conservation of a phenylalanine129 substitution in all CYP51 F5s analyzed. A high resolution X-ray crystal structure of Saccharomyces cerevisiae LDM in complex with VCZ was used for generating a homology model of R. arrhizus CYP51 F5. Structural and functional knowledge of S. cerevisiae CYP51 shows that the F129 residue in Mucorales CYP51 F5 is responsible for intrinsic resistance of Mucorales against short-tailed triazoles, with a V to A substitution in Helix I also potentially playing a role.
Conflict of interest statement
C.L.-F. received research grants, travel grants or honorarium as a speaker or advisor from Gilead Sciences, Pfizer, Schering Plough, MSD and Basilea. M.L. received research grants, travel grants and honorarium as a speaker from MSD, Astellas, BD and Basilea. R.C. received one travel grant from Astellas. The authors T.L., B.M. and J.T. have no potential conflicts of interests to declare.
Figures

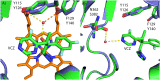
Similar articles
-
Heterologous Expression of Full-Length Lanosterol 14α-Demethylases of Prominent Fungal Pathogens Candida albicans and Candida glabrata Provides Tools for Antifungal Discovery.Antimicrob Agents Chemother. 2018 Oct 24;62(11):e01131-18. doi: 10.1128/AAC.01131-18. Print 2018 Nov. Antimicrob Agents Chemother. 2018. PMID: 30126959 Free PMC article.
-
Impact of Homologous Resistance Mutations from Pathogenic Yeast on Saccharomyces cerevisiae Lanosterol 14α-Demethylase.Antimicrob Agents Chemother. 2018 Feb 23;62(3):e02242-17. doi: 10.1128/AAC.02242-17. Print 2018 Mar. Antimicrob Agents Chemother. 2018. PMID: 29263059 Free PMC article.
-
Characterization of the sterol 14α-demethylases of Fusarium graminearum identifies a novel genus-specific CYP51 function.New Phytol. 2013 May;198(3):821-835. doi: 10.1111/nph.12193. Epub 2013 Feb 27. New Phytol. 2013. PMID: 23442154
-
Fungal cytochrome P450 sterol 14α-demethylase (CYP51) and azole resistance in plant and human pathogens.Appl Microbiol Biotechnol. 2012 Aug;95(4):825-40. doi: 10.1007/s00253-012-4195-9. Epub 2012 Jun 12. Appl Microbiol Biotechnol. 2012. PMID: 22684327 Review.
-
Cytochrome P450 lanosterol 14α-demethylase (CYP51): insights from molecular genetic analysis of the ERG11 gene in Saccharomyces cerevisiae.J Steroid Biochem Mol Biol. 1992 Dec;43(8):1107-16. doi: 10.1016/0960-0760(92)90339-K. J Steroid Biochem Mol Biol. 1992. PMID: 22217856 Review.
Cited by
-
Mucorales Species and Macrophages.J Fungi (Basel). 2020 Jun 26;6(2):94. doi: 10.3390/jof6020094. J Fungi (Basel). 2020. PMID: 32604972 Free PMC article. Review.
-
Characterization of the Sterol 24-C-Methyltransferase Genes Reveals a Network of Alternative Sterol Biosynthetic Pathways in Mucor lusitanicus.Microbiol Spectr. 2023 Jun 15;11(3):e0031523. doi: 10.1128/spectrum.00315-23. Epub 2023 Apr 10. Microbiol Spectr. 2023. PMID: 37036336 Free PMC article.
-
Molecular mechanisms that govern infection and antifungal resistance in Mucorales.Microbiol Mol Biol Rev. 2024 Mar 27;88(1):e0018822. doi: 10.1128/mmbr.00188-22. Epub 2024 Mar 6. Microbiol Mol Biol Rev. 2024. PMID: 38445820 Free PMC article. Review.
-
Mucor circinelloides Thrives inside the Phagosome through an Atf-Mediated Germination Pathway.mBio. 2019 Feb 5;10(1):e02765-18. doi: 10.1128/mBio.02765-18. mBio. 2019. PMID: 30723131 Free PMC article.
-
Antifungal-resistant Mucorales in different indoor environments.Mycology. 2018 Nov 26;10(2):75-83. doi: 10.1080/21501203.2018.1551251. eCollection 2019 Jun. Mycology. 2018. PMID: 31069121 Free PMC article.
References
-
- De Hoog, G. S., Guarro, J., Gené, J. & Figueiras, M. J. Atlas of clinical fungi, second edition. 125–225 (American Society for Microbiology, 2000).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

