Overexpression of suppressive microRNAs, miR-30a and miR-200c are associated with improved survival of breast cancer patients
- PMID: 29162923
- PMCID: PMC5698306
- DOI: 10.1038/s41598-017-16112-y
Overexpression of suppressive microRNAs, miR-30a and miR-200c are associated with improved survival of breast cancer patients
Abstract
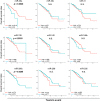
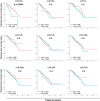
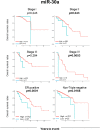
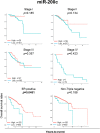
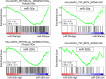
Some microRNAs (miRNAs) are known to suppress breast cancer. However, whether the expressions of these tumor suppressive miRNAs translate to patient survival were not investigated in large cohort. Nine miRNAs (miR-30a, miR-30c, miR-31, miR-126, miR-140, miR-146b, miR-200c, miR-206, and miR-335) known to be tumor suppressive miRNAs in breast cancer were investigated in Genomic Data Common data portal miRNA-Seq dataset and The Cancer Genome Atlas (TCGA) (n = 1052). Of the 9 miRNAs, miR-30a, miR-30c, miR-126, miR-140, miR-206, and miR-335 were found to have significantly lower expression in breast cancer tissues compared to paired normal breast tissue. High expression of miR-30a or miR-200c was associated with significantly better overall survival (OS). Gene Set Enrichment Analysis (GSEA) demonstrated that low expression levels of miR-30a had the tendency to associate with gene enrichment of EMT, while miR-200c did not, in TCGA cohort, and our findings support the need of validation using large cohort to use miRNA as prognostic biomarker for patients with breast cancer.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

