Iron Acquisition Mechanisms and Their Role in the Virulence of Burkholderia Species
- PMID: 29164069
- PMCID: PMC5681537
- DOI: 10.3389/fcimb.2017.00460
Iron Acquisition Mechanisms and Their Role in the Virulence of Burkholderia Species
Erratum in
-
Corrigendum: Iron Acquisition Mechanisms and Their Role in the Virulence of Burkholderia Species.Front Cell Infect Microbiol. 2018 Aug 24;8:305. doi: 10.3389/fcimb.2018.00305. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30181989 Free PMC article.
Abstract
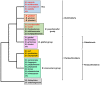
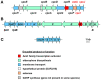
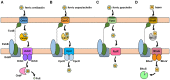
Burkholderia is a genus within the β-Proteobacteriaceae that contains at least 90 validly named species which can be found in a diverse range of environments. A number of pathogenic species occur within the genus. These include Burkholderia cenocepacia and Burkholderia multivorans, opportunistic pathogens that can infect the lungs of patients with cystic fibrosis, and are members of the Burkholderia cepacia complex (Bcc). Burkholderia pseudomallei is also an opportunistic pathogen, but in contrast to Bcc species it causes the tropical human disease melioidosis, while its close relative Burkholderia mallei is the causative agent of glanders in horses. For these pathogens to survive within a host and cause disease they must be able to acquire iron. This chemical element is essential for nearly all living organisms due to its important role in many enzymes and metabolic processes. In the mammalian host, the amount of accessible free iron is negligible due to the low solubility of the metal ion in its higher oxidation state and the tight binding of this element by host proteins such as ferritin and lactoferrin. As with other pathogenic bacteria, Burkholderia species have evolved an array of iron acquisition mechanisms with which to capture iron from the host environment. These mechanisms include the production and utilization of siderophores and the possession of a haem uptake system. Here, we summarize the known mechanisms of iron acquisition in pathogenic Burkholderia species and discuss the evidence for their importance in the context of virulence and the establishment of infection in the host. We have also carried out an extensive bioinformatic analysis to identify which siderophores are produced by each Burkholderia species that is pathogenic to humans.
Keywords: Burkholderia; cystic fibrosis; haem uptake; iron; melioidosis; siderophores.
Figures


References
-
- Agnoli K., Lowe C. A., Farmer K. L., Husnain S. I., Thomas M. S. (2006). The ornibactin biosynthesis and transport genes of Burkholderia cenocepacia are regulated by an extracytoplasmic function sigma factor which is a part of the Fur regulon. J. Bacteriol. 188, 3631–3644. 10.1128/JB.188.10.3631-3644.2006 - DOI - PMC - PubMed
-
- Alice A. F., Lopez C. S., Lowe C. A., Ledesma M. A., Crosa J. H. (2006). Genetic and transcriptional analysis of the siderophore malleobactin biosynthesis and transport genes in the human pathogen Burkholderia pseudomallei K96243. J. Bacteriol. 188, 1551–1566. 10.1128/JB.188.4.1551-1566.2006 - DOI - PMC - PubMed
-
- Amornrit W. M. V., Wangteeraprasert T., Korbsrisate S. (2012). Elevated intracellular levels of iron in host cells promotes Burkholderia pseudomallei infection. Asian Biomed. 6, 465–471. 10.5372/1905-7415.0603.078 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

