Expression of Mitochondrial-Encoded Genes in Blood Differentiate Acute Renal Allograft Rejection
- PMID: 29164120
- PMCID: PMC5671971
- DOI: 10.3389/fmed.2017.00185
Expression of Mitochondrial-Encoded Genes in Blood Differentiate Acute Renal Allograft Rejection
Abstract
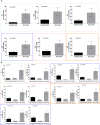
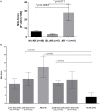
Despite potent immunosuppression, clinical and biopsy confirmed acute renal allograft rejection (AR) still occurs in 10-15% of recipients, ~30% of patients demonstrate subclinical rejection on biopsy, and ~50% of them can show molecular inflammation, all which increase the risk of chronic dysfunction and worsened allograft outcomes. Mitochondria represent intracellular endogenous triggers of inflammation, which can regulate immune cell differentiation, and expansion and cause antigen-independent graft injury, potentially enhancing the development of acute rejection. In the present study, we investigated the role of mitochondrial DNA encoded gene expression in biopsy matched peripheral blood (PB) samples from kidney transplant recipients. Quantitative PCR was performed in 155 PB samples from 115 unique pediatric (<21 years) and adult (>21 years) renal allograft recipients at the point of AR (n = 61) and absence of rejection (n = 94) for the expression of 11 mitochondrial DNA encoded genes. We observed increased expression of all genes in adult recipients compared to pediatric recipients; separate analyses in both cohorts demonstrated increased expression during rejection, which also differentiated borderline rejection and showed an increasing pattern in serially collected samples (0-3 months prior to and post rejection). Our results provide new insights on the role of mitochondria during rejection and potentially indicate mitochondria as targets for novel immunosuppression.
Keywords: acute rejection; biomarkers; gene expression analysis; kidney transplantation; mitochondria.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

