The Sec System: Protein Export in Escherichia coli
- PMID: 29165233
- PMCID: PMC5807066
- DOI: 10.1128/ecosalplus.ESP-0002-2017
The Sec System: Protein Export in Escherichia coli
Abstract
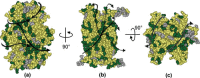
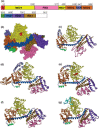
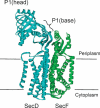
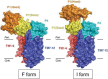
In Escherichia coli, proteins found in the periplasm or the outer membrane are exported from the cytoplasm by the general secretory, Sec, system before they acquire stably folded structure. This dynamic process involves intricate interactions among cytoplasmic and membrane proteins, both peripheral and integral, as well as lipids. In vivo, both ATP hydrolysis and proton motive force are required. Here, we review the Sec system from the inception of the field through early 2016, including biochemical, genetic, and structural data.
Figures

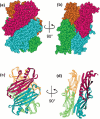
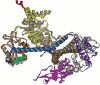
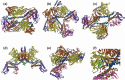
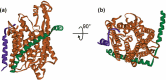
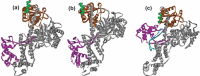
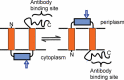
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

