The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: implications on transcription
- PMID: 29165690
- PMCID: PMC5778462
- DOI: 10.1093/nar/gkx1142
The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: implications on transcription
Abstract
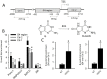
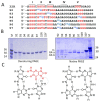
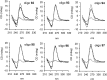
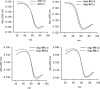
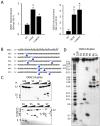
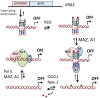
KRAS is one of the most mutated genes in human cancer. It is controlled by a G4 motif located upstream of the transcription start site. In this paper, we demonstrate that 8-oxoguanine (8-oxoG), being more abundant in G4 than in non-G4 regions, is a new player in the regulation of this oncogene. We designed oligonucleotides mimicking the KRAS G4-motif and found that 8-oxoG impacts folding and stability of the G-quadruplex. Dimethylsulphate-footprinting showed that the G-run carrying 8-oxoG is excluded from the G-tetrads and replaced by a redundant G-run in the KRAS G4-motif. Chromatin immunoprecipitation revealed that the base-excision repair protein OGG1 is recruited to the KRAS promoter when the level of 8-oxoG in the G4 region is raised by H2O2. Polyacrylamide gel electrophoresis evidenced that OGG1 removes 8-oxoG from the G4-motif in duplex, but when folded it binds to the G-quadruplex in a non-productive way. We also found that 8-oxoG enhances the recruitment to the KRAS promoter of MAZ and hnRNP A1, two nuclear factors essential for transcription. All this suggests that 8-oxoG in the promoter G4 region could have an epigenetic potential for the control of gene expression.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Sosa V., Moliné T., Somoza S., Paciucci R., Kondoh H.. Oxidative stress and cancer: an overview. Ageing Res. Rev. 2012; 12:376–390. - PubMed
-
- Giorgio M., Trinei M., Migliaccio E., Pelicci P.G.. Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals. Nat. Rev. Mol. Cell Biol. 2007; 8:722–728. - PubMed
-
- St-Pierre J., Buckingham J., Roebuck S.J., Brand M.D.. Topology of superoxide production from different sites in mitochondrial electron transport chain. J. Biol. Chem. 2002; 277:44784–44790. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

