Spatiotemporal Proteomic Profiling of Huntington's Disease Inclusions Reveals Widespread Loss of Protein Function
- PMID: 29166617
- PMCID: PMC5714591
- DOI: 10.1016/j.celrep.2017.10.097
Spatiotemporal Proteomic Profiling of Huntington's Disease Inclusions Reveals Widespread Loss of Protein Function
Abstract
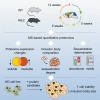
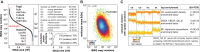
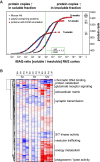
Aggregation of polyglutamine-expanded huntingtin exon 1 (HttEx1) in Huntington's disease (HD) proceeds from soluble oligomers to late-stage inclusions. The nature of the aggregates and how they lead to neuronal dysfunction is not well understood. We employed mass spectrometry (MS)-based quantitative proteomics to dissect spatiotemporal mechanisms of neurodegeneration using the R6/2 mouse model of HD. Extensive remodeling of the soluble brain proteome correlated with insoluble aggregate formation during disease progression. In-depth and quantitative characterization of the aggregates uncovered an unprecedented complexity of several hundred proteins. Sequestration to aggregates depended on protein expression levels and sequence features such as low-complexity regions or coiled-coil domains. In a cell-based HD model, overexpression of a subset of the sequestered proteins in most cases rescued viability and reduced aggregate size. Our spatiotemporally resolved proteome resource of HD progression indicates that widespread loss of cellular protein function contributes to aggregate-mediated toxicity.
Keywords: Huntington’s disease; cerebrospinal fluid; inclusion bodies; neurodegeneration; quantitative proteomics.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Aebersold R., Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. - PubMed
-
- Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. - PubMed
-
- Baldo B., Paganetti P., Grueninger S., Marcellin D., Kaltenbach L.S., Lo D.C., Semmelroth M., Zivanovic A., Abramowski D., Smith D. TR-FRET-based duplex immunoassay reveals an inverse correlation of soluble and aggregated mutant huntingtin in Huntington’s disease. Chem. Biol. 2012;19:264–275. - PubMed
-
- Byrne L.M., Wild E.J. Cerebrospinal fluid biomarkers for Huntington’s disease. J. Huntingtons Dis. 2016;5:1–13. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

