Characterization of the inner membrane protein BB0173 from Borrelia burgdorferi
- PMID: 29166863
- PMCID: PMC5700661
- DOI: 10.1186/s12866-017-1127-y
Characterization of the inner membrane protein BB0173 from Borrelia burgdorferi
Abstract
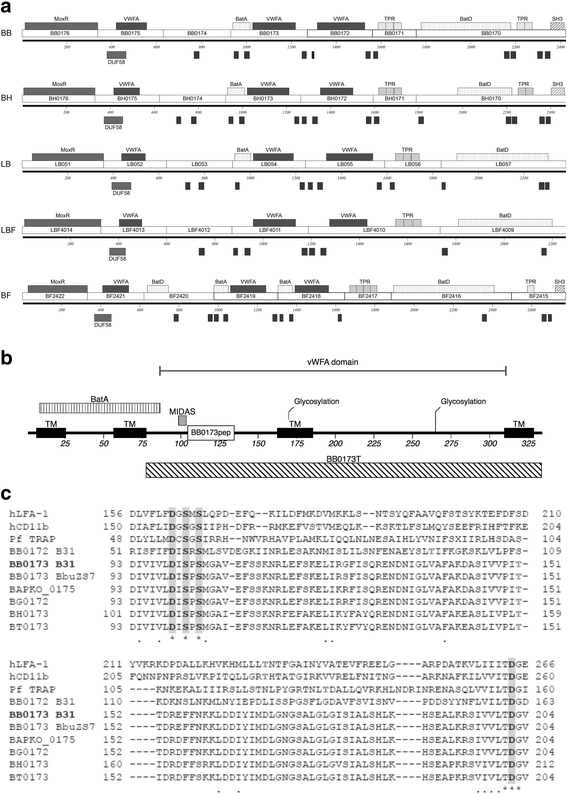
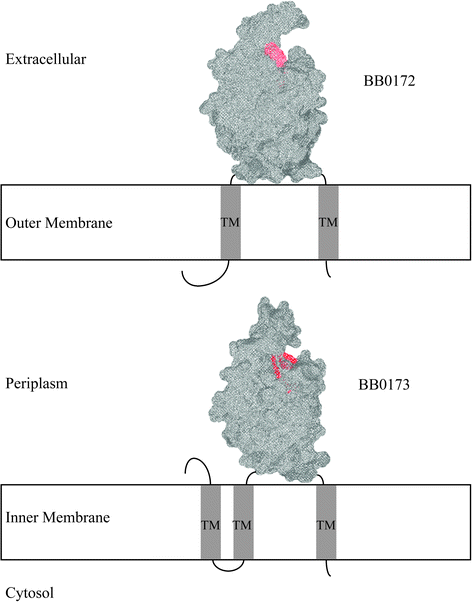
Background: The bacterial spirochete Borrelia burgdorferi is the causative agent of the most commonly reported arthropod-borne illness in the United States, Lyme disease. A family of proteins containing von Willebrand Factor A (VWFA) domains adjacent to a MoxR AAA+ ATPase have been found to be highly conserved in the genus Borrelia. Previously, a VWFA domain containing protein of B. burgdorferi, BB0172, was determined to be an outer membrane protein capable of binding integrin α3β1. In this study, the characterization of a new VWFA domain containing membrane protein, BB0173, is evaluated in order to define the location and topology of this multi-spanning membrane protein. In addition, functional predictions are made.
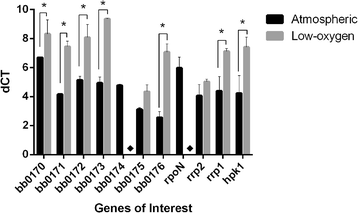
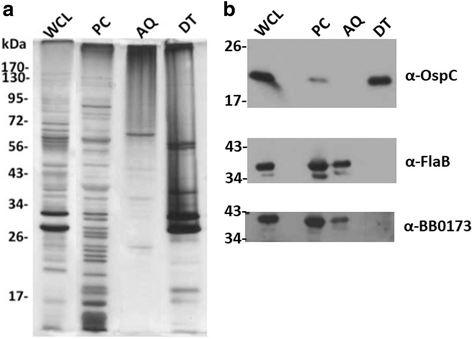
Results: Our results show that BB0173, in contrast to BB0172, is an inner membrane protein, in which the VWFA domain is exposed to the periplasmic space. Further, BB0173 was predicted to have an aerotolerance regulator domain, and expression of BB0173 and the surrounding genes was evaluated under aerobic and microaerophilic conditions, revealing that these genes are downregulated under aerobic conditions. Since the VWFA domain containing proteins of B. burgdorferi are highly conserved, they are likely required for survival of the pathogen through sensing diverse environmental oxygen conditions.
Conclusions: Presently, the complex mechanisms that B. burgdorferi uses to detect and respond to environmental changes are not completely understood. However, studying the mechanisms that allow B. burgdorferi to survive in the highly disparate environments of the tick vector and mammalian host could allow for the development of novel methods of preventing acquisition, survival, or transmission of the spirochete. In this regard, a putative membrane protein, BB0173, was characterized. BB0173 was found to be highly conserved across pathogenic Borrelia, and additionally contains several truly transmembrane domains, and a Bacteroides aerotolerance-like domain. The presence of these functional domains and the highly conserved nature of this protein, strongly suggests a required function of BB0173 in the survival of B. burgdorferi.
Keywords: Aerotolerance; Borrelia burgdorferi; MIDAS motif; Transmembrane; vonWillebrand factor a.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
-
- Centers for Disease C Final 2014 reports of nationally notifiable infectious diseases. MMWR Morb Mortal Wkly Rep. 2015;64(36):1019–1033. doi: 10.15585/mmwr.mm6436a8. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

