Ring finger protein 126 (RNF126) suppresses ionizing radiation-induced p53-binding protein 1 (53BP1) focus formation
- PMID: 29167269
- PMCID: PMC5767864
- DOI: 10.1074/jbc.M116.765602
Ring finger protein 126 (RNF126) suppresses ionizing radiation-induced p53-binding protein 1 (53BP1) focus formation
Abstract
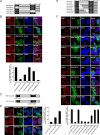
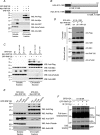
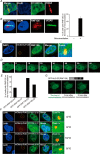
Cells have evolved sophisticated mechanisms to maintain genomic integrity in response to DNA damage. Ionizing radiation (IR)-induced DNA damage results in the formation of IR-induced foci (iRIF) in the nucleus. The iRIF formation is part of the DNA damage response (DDR), which is an essential signaling cascade that must be strictly regulated because either the loss of or an augmented DDR leads to loss of genome integrity. Accordingly, negative regulation of the DDR is as critical as its activation. In this study, we have identified ring finger protein 126 (RNF126) as a negative regulator of the DDR from a screen of iRIF containing 53BP1. RNF126 overexpression abolishes not only the formation of 53BP1 iRIF but also of RNF168, FK2, RAP80, and BRCA1. However, the iRIF formation of γH2AX, MDC1, and RNF8 is maintained, indicating that RNF126 acts between RNF8 and RNF168 during the DDR. In addition, RNF126 overexpression consistently results in the loss of RNF168-mediated H2A monoubiquitination at lysine 13/15 and inhibition of the non-homologous end joining capability. Taken together, our findings reveal that RNF126 is a novel factor involved in the negative regulation of DDR, which is important for sustaining genomic integrity.
Keywords: DNA damage; DNA damage response; histone modification; signal transduction; ubiquitylation (ubiquitination).
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

