Evaluation of Whole-Genome Sequencing for Mycobacterial Species Identification and Drug Susceptibility Testing in a Clinical Setting: a Large-Scale Prospective Assessment of Performance against Line Probe Assays and Phenotyping
- PMID: 29167290
- PMCID: PMC5786738
- DOI: 10.1128/JCM.01480-17
Evaluation of Whole-Genome Sequencing for Mycobacterial Species Identification and Drug Susceptibility Testing in a Clinical Setting: a Large-Scale Prospective Assessment of Performance against Line Probe Assays and Phenotyping
Abstract
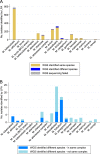
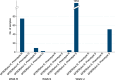
Use of whole-genome sequencing (WGS) for routine mycobacterial species identification and drug susceptibility testing (DST) is becoming a reality. We compared the performances of WGS and standard laboratory workflows prospectively, by parallel processing at a major mycobacterial reference service over the course of 1 year, for species identification, first-line Mycobacterium tuberculosis resistance prediction, and turnaround time. Among 2,039 isolates with line probe assay results for species identification, 74 (3.6%) failed sequencing or WGS species identification. Excluding these isolates, clinically important species were identified for 1,902 isolates, of which 1,825 (96.0%) were identified as the same species by WGS and the line probe assay. A total of 2,157 line probe test results for detection of resistance to the first-line drugs isoniazid and rifampin were available for 728 M. tuberculosis complex isolates. Excluding 216 (10.0%) cases where there were insufficient sequencing data for WGS to make a prediction, overall concordance was 99.3% (95% confidence interval [CI], 98.9 to 99.6%), sensitivity was 97.6% (91.7 to 99.7%), and specificity was 99.5% (99.0 to 99.7%). A total of 2,982 phenotypic DST results were available for 777 M. tuberculosis complex isolates. Of these, 356 (11.9%) had no WGS comparator due to insufficient sequencing data, and in 154 (5.2%) cases the WGS prediction was indeterminate due to discovery of novel, previously uncharacterized mutations. Excluding these data, overall concordance was 99.2% (98.7 to 99.5%), sensitivity was 94.2% (88.4 to 97.6%), and specificity was 99.4% (99.0 to 99.7%). Median processing times for the routine laboratory tests versus WGS were similar overall, i.e., 20 days (interquartile range [IQR], 15 to 31 days) and 21 days (15 to 29 days), respectively (P = 0.41). In conclusion, WGS predicts species and drug susceptibility with great accuracy, but work is needed to increase the proportion of predictions made.
Keywords: WGS; line probe assay; mycobacteria; phenotype; whole-genome sequencing.
Copyright © 2018 Quan et al.
Figures


References
-
- World Health Organization. 2016. Global tuberculosis report 2016. WHO, Geneva, Switzerland.
-
- Nathavitharana RR, Cudahy PG, Schumacher SG, Steingart KR, Pai M, Denkinger CM. 2017. Accuracy of line probe assays for the diagnosis of pulmonary and multidrug-resistant tuberculosis: a systematic review and meta-analysis. Eur Respir J 49:1601075. doi:10.1183/13993003.01075-2016. - DOI - PMC - PubMed
-
- Nathavitharana RR, Hillemann D, Schumacher SG, Schlueter B, Ismail N, Omar SV, Sikhondze W, Havumaki J, Valli E, Boehme C, Denkinger CM. 2016. Multicenter noninferiority evaluation of Hain GenoType MTBDRplus version 2 and Nipro NTM+MDRTB line probe assays for detection of rifampin and isoniazid resistance. J Clin Microbiol 54:1624–1630. doi:10.1128/JCM.00251-16. - DOI - PMC - PubMed
-
- Tortoli E, Richter E, Borroni E, Cabibbe AM, Capitolo E, Cittaro D, Engel R, Hendricks O, Hillemann D, Kristiansen JE, Mariottini A, Schubert S, Cirillo DM. 2016. Mycobacterium alsense sp. nov., a scotochromogenic slow grower isolated from clinical respiratory specimens. Int J Syst Evol Microbiol 66:450–456. doi:10.1099/ijsem.0.000744. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

