Differential Immunodominance Hierarchy of CD8+ T-Cell Responses in HLA-B*27:05- and -B*27:02-Mediated Control of HIV-1 Infection
- PMID: 29167337
- PMCID: PMC5790925
- DOI: 10.1128/JVI.01685-17
Differential Immunodominance Hierarchy of CD8+ T-Cell Responses in HLA-B*27:05- and -B*27:02-Mediated Control of HIV-1 Infection
Abstract
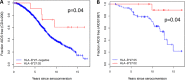
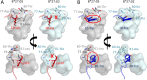
The well-characterized association between HLA-B*27:05 and protection against HIV disease progression has been linked to immunodominant HLA-B*27:05-restricted CD8+ T-cell responses toward the conserved Gag KK10 (residues 263 to 272) and polymerase (Pol) KY9 (residues 901 to 909) epitopes. We studied the impact of the 3 amino acid differences between HLA-B*27:05 and the closely related HLA-B*27:02 on the HIV-specific CD8+ T-cell response hierarchy and on immune control of HIV. Genetic epidemiological data indicate that both HLA-B*27:02 and HLA-B*27:05 are associated with slower disease progression and lower viral loads. The effect of HLA-B*27:02 appeared to be consistently stronger than that of HLA-B*27:05. In contrast to HLA-B*27:05, the immunodominant HIV-specific HLA-B*27:02-restricted CD8+ T-cell response is to a Nef epitope (residues 142 to 150 [VW9]), with Pol KY9 subdominant and Gag KK10 further subdominant. This selection was driven by structural differences in the F pocket, mediated by a polymorphism between these two HLA alleles at position 81. Analysis of autologous virus sequences showed that in HLA-B*27:02-positive subjects, all three of these CD8+ T-cell responses impose selection pressure on the virus, whereas in HLA-B*27:05-positive subjects, there is no Nef VW9-mediated selection pressure. These studies demonstrate that HLA-B*27:02 mediates protection against HIV disease progression that is at least as strong as or stronger than that mediated by HLA-B*27:05. In combination with the protective Gag KK10 and Pol KY9 CD8+ T-cell responses that dominate HIV-specific CD8+ T-cell activity in HLA-B*27:05-positive subjects, a Nef VW9-specific response is additionally present and immunodominant in HLA-B*27:02-positive subjects, mediated through a polymorphism at residue 81 in the F pocket, that contributes to selection pressure against HIV.IMPORTANCE CD8+ T cells play a central role in successful control of HIV infection and have the potential also to mediate the eradication of viral reservoirs of infection. The principal means by which protective HLA class I molecules, such as HLA-B*27:05 and HLA-B*57:01, slow HIV disease progression is believed to be via the particular HIV-specific CD8+ T cell responses restricted by those alleles. We focus here on HLA-B*27:05, one of the best-characterized protective HLA molecules, and the closely related HLA-B*27:02, which differs by only 3 amino acids and which has not been well studied in relation to control of HIV infection. We show that HLA-B*27:02 is also protective against HIV disease progression, but the CD8+ T-cell immunodominance hierarchy of HLA-B*27:02 differs strikingly from that of HLA-B*27:05. These findings indicate that the immunodominant HLA-B*27:02-restricted Nef response adds to protection mediated by the Gag and Pol specificities that dominate anti-HIV CD8+ T-cell activity in HLA-B*27:05-positive subjects.
Keywords: CD8+ T cell; HIV Gag; HIV Nef; HLA; HLA-B*27; human immunodeficiency virus.
Copyright © 2018 Adland et al.
Figures


References
-
- Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, Cirulli ET, Urban TJ, Zhang K, Gumbs CE, Smith JP, Castagna A, Cozzi-Lepri A, De Luca A, Easterbrook P, Gunthard HF, Mallal S, Mussini C, Dalmau J, Martinez-Picado J, Miro JM, Obel N, Wolinsky SM, Martinson JJ, Detels R, Margolick JB, Jacobson LP, Descombes P, Antonarakis SE, Beckmann JS, O'Brien SJ, Letvin NL, McMichael AJ, Haynes BF, Carrington M, Feng S, Telenti A, Goldstein DB. 2009. Common genetic variation and the control of HIV-1 in humans. PLoS Genet 5:e1000791. doi: 10.1371/journal.pgen.1000791. - DOI - PMC - PubMed
-
- Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O'Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DM, Vine S, Addo MM, et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557. doi: 10.1126/science.1195271. - DOI - PMC - PubMed
-
- Kaslow RA, Carrington M, Apple R, Park L, Munoz A, Saah AJ, Goedert JJ, Winkler C, O'Brien SJ, Rinaldo C, Detels R, Blattner W, Phair J, Erlich H, Mann DL. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med 2:405–411. doi: 10.1038/nm0496-405. - DOI - PubMed
-
- Madden DR, Gorga JC, Strominger JL, Wiley DC. 1992. The three-dimensional structure of HLA-B27 at 2.1 A resolution suggests a general mechanism for tight peptide binding to MHC. Cell 70:1035–1048. - PubMed
-
- Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael AJ, Rowland-Jones S. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med 3:212–217. doi: 10.1038/nm0297-212. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 AI035042/AI/NIAID NIH HHS/United States
- MR/K012037/1/MRC_/Medical Research Council/United Kingdom
- UM1 AI035043/AI/NIAID NIH HHS/United States
- MR/L006588/1/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- U01 AI035040/AI/NIAID NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- R01 AI046995/AI/NIAID NIH HHS/United States
- U01 AI035039/AI/NIAID NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- MR/K012037/MRC_/Medical Research Council/United Kingdom
- U19 AI096109/AI/NIAID NIH HHS/United States
- U01 AI035041/AI/NIAID NIH HHS/United States
- R24 AI067039/AI/NIAID NIH HHS/United States
- WT104748MA/WT_/Wellcome Trust/United Kingdom
- P30 AI027763/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

