Transcriptome and metabolite analyses reveal the complex metabolic genes involved in volatile terpenoid biosynthesis in garden sage (Salvia officinalis)
- PMID: 29167468
- PMCID: PMC5700130
- DOI: 10.1038/s41598-017-15478-3
Transcriptome and metabolite analyses reveal the complex metabolic genes involved in volatile terpenoid biosynthesis in garden sage (Salvia officinalis)
Abstract
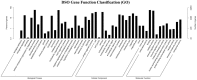
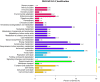
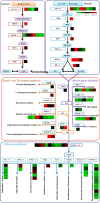
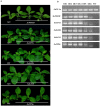
A large number of terpenoid compounds have been extracted from different tissues of S. officinalis. However, the molecular genetic basis of terpene biosynthesis pathways is virtually unknown. In this study, approximately 6.6 Gb of raw data were generated from the transcriptome of S. officinalis leaves using Illumina HiSeq 2000 sequencing. After filtering and removing the adapter sequences from the raw data, the number of reads reached 21 million, comprising 98 million of high-quality nucleotide bases. 48,671 unigenes were assembled de novo and annotated for establishing a valid database for studying terpenoid biosynthesis. We identified 135 unigenes that are putatively involved in terpenoid metabolism, including 70 mevalonate and methyl-erythritol phosphate pathways, terpenoid backbone biosynthesis genes, and 65 terpene synthase genes. Moreover, five terpene synthase genes were studied for their functions in terpenoid biosynthesis by using transgenic tobacco; most transgenic tobacco plants expressing these terpene synthetic genes produced increased amounts of terpenoids compared with wild-type control. The combined data analyses from the transcriptome and metabolome provide new insights into our understanding of the complex metabolic genes in terpenoid-rich sage, and our study paves the way for the future metabolic engineering of the biosynthesis of useful terpene compounds in S. officinalis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Alziar, G. Catalogue synonymique des Salvia L. dumonde (Lamiaceae). 5 (3–4):87–136; 6(1–2, 4):79–115, 163–204; 7(1–2):59–109; 9(2–3):413–497; 10(3–4). (I.–VI. Biocosme Mesogeén) 33–117 (1988–1993).
-
- Carretero-Paulet L, et al. Campositionm Expression and molecular analysis of the Arabidopsis DXR gene encoding 1-Deoxy-D-xylulose-5-phosphate reductoisomerase, the first committed enzyme of the 2-C-Methyl-D-erythritol-4-phosphate pathway. Plant Physiol. 2002;129:1581–1591. doi: 10.1104/pp.003798. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

