Gene Editing and Crop Improvement Using CRISPR-Cas9 System
- PMID: 29167680
- PMCID: PMC5682324
- DOI: 10.3389/fpls.2017.01932
Gene Editing and Crop Improvement Using CRISPR-Cas9 System
Abstract
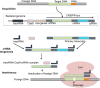
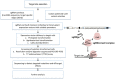
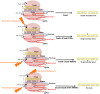
Advancements in Genome editing technologies have revolutionized the fields of functional genomics and crop improvement. CRISPR/Cas9 (clustered regularly interspaced short palindromic repeat)-Cas9 is a multipurpose technology for genetic engineering that relies on the complementarity of the guideRNA (gRNA) to a specific sequence and the Cas9 endonuclease activity. It has broadened the agricultural research area, bringing in new opportunities to develop novel plant varieties with deletion of detrimental traits or addition of significant characters. This RNA guided genome editing technology is turning out to be a groundbreaking innovation in distinct branches of plant biology. CRISPR technology is constantly advancing including options for various genetic manipulations like generating knockouts; making precise modifications, multiplex genome engineering, and activation and repression of target genes. The review highlights the progression throughout the CRISPR legacy. We have studied the rapid evolution of CRISPR/Cas9 tools with myriad functionalities, capabilities, and specialized applications. Among varied diligences, plant nutritional improvement, enhancement of plant disease resistance and production of drought tolerant plants are reviewed. The review also includes some information on traditional delivery methods of Cas9-gRNA complexes into plant cells and incorporates the advent of CRISPR ribonucleoproteins (RNPs) that came up as a solution to various limitations that prevailed with plasmid-based CRISPR system.
Keywords: CRISPR ribonucleoproteins; CRISPR/Cas system; disease resistance; gene expression regulation; genome editing; metabolic engineering; nutrition improvement.
Figures



References
-
- Ahloowalia B. S., Maluszynski M. (2001). Induced mutations- A new paradigm in plant breeding. Euphytica 118 167–173. 10.1023/A:1004162323428 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

