Primer in Genetics and Genomics, Article 6: Basics of Epigenetic Control
- PMID: 29168388
- PMCID: PMC6949967
- DOI: 10.1177/1099800417742967
Primer in Genetics and Genomics, Article 6: Basics of Epigenetic Control
Abstract
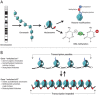
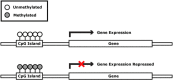
The epigenome is a collection of chemical compounds that attach to and overlay the DNA sequence to direct gene expression. Epigenetic marks do not alter DNA sequence but instead allow or silence gene activity and the subsequent production of proteins that guide the growth and development of an organism, direct and maintain cell identity, and allow for the production of primordial germ cells (PGCs; ova and spermatozoa). The three main epigenetic marks are (1) histone modification, (2) DNA methylation, and (3) noncoding RNA, and each works in a different way to regulate gene expression. This article reviews these concepts and discusses their role in normal functions such as X-chromosome inactivation, epigenetic reprogramming during embryonic development and PGC production, and the clinical example of the imprinting disorders Angelman and Prader-Willi syndromes.
Keywords: DNA methylation; X-chromosome inactivation; chromatin; epigenomics; histone code.
Conflict of interest statement
Figures

Similar articles
-
Promoter DNA methylation couples genome-defence mechanisms to epigenetic reprogramming in the mouse germline.Development. 2012 Oct;139(19):3623-32. doi: 10.1242/dev.081661. Development. 2012. PMID: 22949617 Free PMC article.
-
[Epigenetic reprogramming, germline and genomic imprinting].Med Sci (Paris). 2024 Dec;40(12):892-903. doi: 10.1051/medsci/2024177. Epub 2024 Dec 20. Med Sci (Paris). 2024. PMID: 39705560 Review. French.
-
Epigenetic profiles in primordial germ cells: global modulation and fine tuning of the epigenome for acquisition of totipotency.Dev Growth Differ. 2010 Aug;52(6):517-25. doi: 10.1111/j.1440-169X.2010.01190.x. Dev Growth Differ. 2010. PMID: 20646024 Review.
-
The influence of DNA methylation on monoallelic expression.Essays Biochem. 2019 Dec 20;63(6):663-676. doi: 10.1042/EBC20190034. Essays Biochem. 2019. PMID: 31782494 Free PMC article. Review.
-
A current view of the epigenome in mouse primordial germ cells.Mol Reprod Dev. 2014 Feb;81(2):160-70. doi: 10.1002/mrd.22214. Epub 2013 Aug 7. Mol Reprod Dev. 2014. PMID: 23868517 Review.
Cited by
-
Transcriptomics and epigenetic data integration learning module on Google Cloud.Brief Bioinform. 2024 Jul 23;25(Supplement_1):bbae352. doi: 10.1093/bib/bbae352. Brief Bioinform. 2024. PMID: 39101486 Free PMC article.
-
Expanding adverse outcome pathways towards one health models for nanosafety.Front Toxicol. 2023 Aug 25;5:1176745. doi: 10.3389/ftox.2023.1176745. eCollection 2023. Front Toxicol. 2023. PMID: 37692900 Free PMC article.
-
Genome‑wide DNA methylation profiling in a rat model with vascular dementia.Mol Med Rep. 2018 Jul;18(1):123-130. doi: 10.3892/mmr.2018.8990. Epub 2018 May 8. Mol Med Rep. 2018. PMID: 29749552 Free PMC article.
-
The Influence of Physical Activity and Epigenomics On Cognitive Function and Brain Health in Breast Cancer.Front Aging Neurosci. 2020 May 8;12:123. doi: 10.3389/fnagi.2020.00123. eCollection 2020. Front Aging Neurosci. 2020. PMID: 32457596 Free PMC article. Review.
-
Using Genetic Burden Scores for Gene-by-Methylation Interaction Analysis on Metabolic Syndrome in African Americans.Biol Res Nurs. 2019 May;21(3):279-285. doi: 10.1177/1099800419828486. Epub 2019 Feb 19. Biol Res Nurs. 2019. PMID: 30781968 Free PMC article.
References
-
- Allis C. D., Jenuwein T. (2016). The molecular hallmarks of epigenetic control. Nature Reviews Genetics, 17, 487–500. - PubMed
-
- Barr M. L., Bertram E. G. (1949). A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature, 163, 676. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

