Global profiling of lysine reactivity and ligandability in the human proteome
- PMID: 29168484
- PMCID: PMC5726523
- DOI: 10.1038/nchem.2826
Global profiling of lysine reactivity and ligandability in the human proteome
Abstract
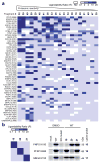
Nucleophilic amino acids make important contributions to protein function, including performing key roles in catalysis and serving as sites for post-translational modification. Electrophilic groups that target amino-acid nucleophiles have been used to create covalent ligands and drugs, but have, so far, been mainly limited to cysteine and serine. Here, we report a chemical proteomic platform for the global and quantitative analysis of lysine residues in native biological systems. We have quantified, in total, more than 9,000 lysines in human cell proteomes and have identified several hundred residues with heightened reactivity that are enriched at protein functional sites and can frequently be targeted by electrophilic small molecules. We have also discovered lysine-reactive fragment electrophiles that inhibit enzymes by active site and allosteric mechanisms, as well as disrupt protein-protein interactions in transcriptional regulatory complexes, emphasizing the broad potential and diverse functional consequences of liganding lysine residues throughout the human proteome.
Conflict of interest statement
The authors declare competing financial interests. Dr. Cravatt is a founder and advisor to Vividion Therapeutics, a biotechnology company interested in using chemical proteomic methods to develop small-molecule drugs to treat human disease.
Figures
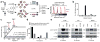
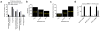
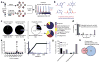


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

