N-terminomics reveals control of Arabidopsis seed storage proteins and proteases by the Arg/N-end rule pathway
- PMID: 29168982
- PMCID: PMC5947142
- DOI: 10.1111/nph.14909
N-terminomics reveals control of Arabidopsis seed storage proteins and proteases by the Arg/N-end rule pathway
Abstract
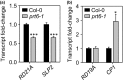
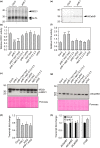
The N-end rule pathway of targeted protein degradation is an important regulator of diverse processes in plants but detailed knowledge regarding its influence on the proteome is lacking. To investigate the impact of the Arg/N-end rule pathway on the proteome of etiolated seedlings, we used terminal amine isotopic labelling of substrates with tandem mass tags (TMT-TAILS) for relative quantification of N-terminal peptides in prt6, an Arabidopsis thaliana N-end rule mutant lacking the E3 ligase PROTEOLYSIS6 (PRT6). TMT-TAILS identified over 4000 unique N-terminal peptides representing c. 2000 protein groups. Forty-five protein groups exhibited significantly increased N-terminal peptide abundance in prt6 seedlings, including cruciferins, major seed storage proteins, which were regulated by Group VII Ethylene Response Factor (ERFVII) transcription factors, known substrates of PRT6. Mobilisation of endosperm α-cruciferin was delayed in prt6 seedlings. N-termini of several proteases were downregulated in prt6, including RD21A. RD21A transcript, protein and activity levels were downregulated in a largely ERFVII-dependent manner. By contrast, cathepsin B3 protein and activity were upregulated by ERFVIIs independent of transcript. We propose that the PRT6 branch of the pathway regulates protease activities in a complex manner and optimises storage reserve mobilisation in the transition from seed to seedling via control of ERFVII action.
Keywords: Arabidopsis thaliana; TAILS; N-end rule; N-terminomics; cruciferin; protease; quantitative proteomics; tandem mass tag (TMT).
© 2017 The Authors. New Phytologist © 2017 New Phytologist Trust.
Figures

Comment in
-
Unravelling the mode of action of plant proteases.New Phytol. 2018 May;218(3):879-881. doi: 10.1111/nph.15156. New Phytol. 2018. PMID: 29658638 No abstract available.
References
-
- Bachmair A, Finley D, Varshavsky A. 1986. In vivo half‐life of a protein is a function of its amino‐terminal residue. Science 234: 179–186. - PubMed
-
- Biniossek ML, Schilling O. 2012. Enhanced identification of peptides lacking basic residues by LC‐ESI‐MS/MS analysis of singly charged peptides. Proteomics 12: 1303–1309. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

