A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation
- PMID: 29170278
- PMCID: PMC5875943
- DOI: 10.1126/science.aan6490
A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation
Abstract
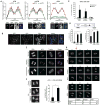
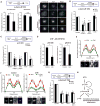
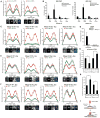
The ataxia telangiectasia mutated and Rad3-related (ATR) kinase is crucial for DNA damage and replication stress responses. Here, we describe an unexpected role of ATR in mitosis. Acute inhibition or degradation of ATR in mitosis induces whole-chromosome missegregation. The effect of ATR ablation is not due to altered cyclin-dependent kinase 1 (CDK1) activity, DNA damage responses, or unscheduled DNA synthesis but to loss of an ATR function at centromeres. In mitosis, ATR localizes to centromeres through Aurora A-regulated association with centromere protein F (CENP-F), allowing ATR to engage replication protein A (RPA)-coated centromeric R loops. As ATR is activated at centromeres, it stimulates Aurora B through Chk1, preventing formation of lagging chromosomes. Thus, a mitosis-specific and R loop-driven ATR pathway acts at centromeres to promote faithful chromosome segregation, revealing functions of R loops and ATR in suppressing chromosome instability.
Copyright © 2018, American Association for the Advancement of Science.
Figures

Comment in
-
A new mitotic activity comes into focus.Science. 2018 Jan 5;359(6371):30-31. doi: 10.1126/science.aar4799. Science. 2018. PMID: 29302000 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

