Methylomic and phenotypic analysis of the ModH5 phasevarion of Helicobacter pylori
- PMID: 29170397
- PMCID: PMC5700931
- DOI: 10.1038/s41598-017-15721-x
Methylomic and phenotypic analysis of the ModH5 phasevarion of Helicobacter pylori
Abstract
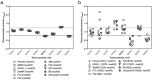
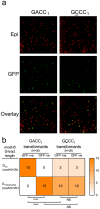
The Helicobacter pylori phase variable gene modH, typified by gene HP1522 in strain 26695, encodes a N6-adenosine type III DNA methyltransferase. Our previous studies identified multiple strain-specific modH variants (modH1 - modH19) and showed that phase variation of modH5 in H. pylori P12 influenced expression of motility-associated genes and outer membrane protein gene hopG. However, the ModH5 DNA recognition motif and the mechanism by which ModH5 controls gene expression were unknown. Here, using comparative single molecule real-time sequencing, we identify the DNA site methylated by ModH5 as 5'-Gm6ACC-3'. This motif is vastly underrepresented in H. pylori genomes, but overrepresented in a number of virulence genes, including motility-associated genes, and outer membrane protein genes. Motility and the number of flagella of H. pylori P12 wild-type were significantly higher than that of isogenic modH5 OFF or ΔmodH5 mutants, indicating that phase variable switching of modH5 expression plays a role in regulating H. pylori motility phenotypes. Using the flagellin A (flaA) gene as a model, we show that ModH5 modulates flaA promoter activity in a GACC methylation-dependent manner. These findings provide novel insights into the role of ModH5 in gene regulation and how it mediates epigenetic regulation of H. pylori motility.
Conflict of interest statement
Y.N.S., R.J.G., P.M.P., K.T., E.L.H., M.P.J. and T.K. declare no competing interests; M.B., T.A.C. and J.K. are full-time employees at Pacific Biosciences, a company developing single-molecule sequencing technologies.
Figures




References
-
- Moxon ER, Thaler DS. Microbial genetics. The tinkerer’s evolving tool-box. Nature. 1997;387:661–662. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

