Characterization of large and small-plaque variants in the Zika virus clinical isolate ZIKV/Hu/S36/Chiba/2016
- PMID: 29170504
- PMCID: PMC5701032
- DOI: 10.1038/s41598-017-16475-2
Characterization of large and small-plaque variants in the Zika virus clinical isolate ZIKV/Hu/S36/Chiba/2016
Abstract
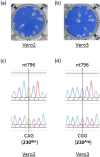
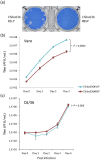
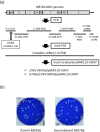
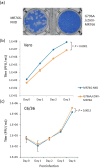
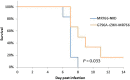
An Asian/American lineage Zika virus (ZIKV) strain ZIKV/Hu/S36/Chiba/2016 formed 2 types in plaque size, large and small. Genomic analysis of the plaque-forming clones obtained from the isolate indicated that the clones forming small plaques commonly had an adenine nucleotide at position 796 (230Gln in the amino acid sequence), while clones forming large plaques had a guanine nucleotide (230Arg) at the same position, suggesting that this position was associated with the difference in plaque size. Growth kinetics of a large-plaque clone was faster than that of a small-plaque clone in Vero cells. Recombinant ZIKV G796A/rZIKV-MR766, which carries a missense G796A mutation, was produced using an infectious molecular clone of the ZIKV MR766 strain rZIKV-MR766/pMW119-CMVP. The plaque size of the G796A mutant was significantly smaller than that of the parental strain. The G796A mutation clearly reduced the growth rate of the parental virus in Vero cells. Furthermore, the G796A mutation also decreased the virulence of the MR766 strain in IFNAR1 knockout mice. These results indicate that the amino acid variation at position 230 in the viral polyprotein, which is located in the M protein sequence, is a molecular determinant for plaque morphology, growth property, and virulence in mice of ZIKV.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Two Genetic Differences between Closely Related Zika Virus Strains Determine Pathogenic Outcome in Mice.J Virol. 2020 Sep 29;94(20):e00618-20. doi: 10.1128/JVI.00618-20. Print 2020 Sep 29. J Virol. 2020. PMID: 32796074 Free PMC article.
-
An Alanine-to-Valine Substitution in the Residue 175 of Zika Virus NS2A Protein Affects Viral RNA Synthesis and Attenuates the Virus In Vivo.Viruses. 2018 Oct 7;10(10):547. doi: 10.3390/v10100547. Viruses. 2018. PMID: 30301244 Free PMC article.
-
Influence of Zika virus 3'-end sequence and nonstructural protein evolution on the viral replication competence and virulence.Emerg Microbes Infect. 2022 Dec;11(1):2447-2465. doi: 10.1080/22221751.2022.2128433. Emerg Microbes Infect. 2022. PMID: 36149812 Free PMC article.
-
Development and Characterization of Recombinant Virus Generated from a New World Zika Virus Infectious Clone.J Virol. 2016 Dec 16;91(1):e01765-16. doi: 10.1128/JVI.01765-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795432 Free PMC article.
-
Novel genetically stable infectious clone for a Zika virus clinical isolate and identification of RNA elements essential for virus production.Virus Res. 2018 Sep 15;257:14-24. doi: 10.1016/j.virusres.2018.08.016. Epub 2018 Aug 23. Virus Res. 2018. PMID: 30144463
Cited by
-
Melatonin attenuates dimethyl sulfoxide- and Zika virus-induced degeneration of porcine induced neural stem cells.In Vitro Cell Dev Biol Anim. 2022 Mar;58(3):232-242. doi: 10.1007/s11626-022-00648-z. Epub 2022 Mar 2. In Vitro Cell Dev Biol Anim. 2022. PMID: 35235152 Free PMC article.
-
Leu-to-Phe substitution at prM146 decreases the growth ability of Zika virus and partially reduces its pathogenicity in mice.Sci Rep. 2021 Oct 4;11(1):19635. doi: 10.1038/s41598-021-99086-2. Sci Rep. 2021. PMID: 34608212 Free PMC article.
-
Insights into In Vitro Adaptation of EV71 and Analysis of Reduced Virulence by In Silico Predictions.Vaccines (Basel). 2023 Mar 11;11(3):629. doi: 10.3390/vaccines11030629. Vaccines (Basel). 2023. PMID: 36992213 Free PMC article.
-
Enhanced Pathogenic Consequences Induced by a Seven-Amino-Acid Extension in the G Protein of the HRSV BA9 Genotype.Int J Mol Sci. 2025 Feb 27;26(5):2081. doi: 10.3390/ijms26052081. Int J Mol Sci. 2025. PMID: 40076707 Free PMC article.
-
The dinucleotide composition of the Zika virus genome is shaped by conflicting evolutionary pressures in mammalian hosts and mosquito vectors.PLoS Biol. 2021 Apr 19;19(4):e3001201. doi: 10.1371/journal.pbio.3001201. eCollection 2021 Apr. PLoS Biol. 2021. PMID: 33872300 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

