A COUP-TFII Human Embryonic Stem Cell Reporter Line to Identify and Select Atrial Cardiomyocytes
- PMID: 29173897
- PMCID: PMC5785710
- DOI: 10.1016/j.stemcr.2017.10.024
A COUP-TFII Human Embryonic Stem Cell Reporter Line to Identify and Select Atrial Cardiomyocytes
Abstract
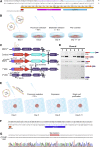
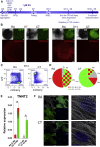
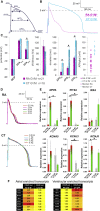
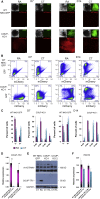
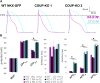
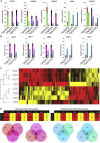
Reporter cell lines have already proven valuable in identifying, tracking, and purifying cardiac subtypes and progenitors during differentiation of human pluripotent stem cells (hPSCs). We previously showed that chick ovalbumin upstream promoter transcription factor II (COUP-TFII) is highly enriched in human atrial cardiomyocytes (CMs), but not ventricular. Here, we targeted mCherry to the COUP-TFII genomic locus in hPSCs expressing GFP from the NKX2.5 locus. This dual atrial NKX2.5EGFP/+-COUP-TFIImCherry/+ reporter line allowed identification and selection of GFP+ (G+)/mCherry+ (M+) CMs following cardiac differentiation. These cells exhibited transcriptional and functional properties of atrial CMs, whereas G+/M- CMs displayed ventricular characteristics. Via CRISPR/Cas9-mediated knockout, we demonstrated that COUP-TFII is not required for atrial specification in hPSCs. This new tool allowed selection of human atrial and ventricular CMs from mixed populations, of relevance for studying cardiac specification, developing human atrial disease models, and examining distinct effects of drugs on the atrium versus ventricle.
Keywords: COUP-TFII-knockout; COUP-TFII-mCherry fluorescent stem cell reporter; CRISPR/Cas9; atrial specification; cardiac differentiation; human embryonic stem cells.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Isolation and characterization of ventricular-like cells derived from NKX2-5eGFP/w and MLC2vmCherry/w double knock-in human pluripotent stem cells.Biochem Biophys Res Commun. 2018 Jan 1;495(1):1278-1284. doi: 10.1016/j.bbrc.2017.11.133. Epub 2017 Nov 22. Biochem Biophys Res Commun. 2018. PMID: 29175323
-
CRISPR/Cas9-based targeting of fluorescent reporters to human iPSCs to isolate atrial and ventricular-specific cardiomyocytes.Sci Rep. 2021 Feb 4;11(1):3026. doi: 10.1038/s41598-021-81860-x. Sci Rep. 2021. PMID: 33542270 Free PMC article.
-
Chemical-defined and albumin-free generation of human atrial and ventricular myocytes from human pluripotent stem cells.Stem Cell Res. 2017 Mar;19:94-103. doi: 10.1016/j.scr.2017.01.006. Epub 2017 Jan 12. Stem Cell Res. 2017. PMID: 28110125
-
Human cardiomyocyte generation from pluripotent stem cells: A state-of-art.Life Sci. 2016 Jan 15;145:98-113. doi: 10.1016/j.lfs.2015.12.023. Epub 2015 Dec 10. Life Sci. 2016. PMID: 26682938 Review.
-
COUP-TFII revisited: Its role in metabolic gene regulation.Steroids. 2019 Jan;141:63-69. doi: 10.1016/j.steroids.2018.11.013. Epub 2018 Nov 24. Steroids. 2019. PMID: 30481528 Free PMC article. Review.
Cited by
-
Improved Atrial Differentiation of Human Pluripotent Stem Cells by Activation of Retinoic Acid Receptor Alpha (RARα).J Pers Med. 2022 Apr 13;12(4):628. doi: 10.3390/jpm12040628. J Pers Med. 2022. PMID: 35455744 Free PMC article.
-
Metabolically driven maturation of human-induced-pluripotent-stem-cell-derived cardiac microtissues on microfluidic chips.Nat Biomed Eng. 2022 Apr;6(4):372-388. doi: 10.1038/s41551-022-00884-4. Epub 2022 Apr 27. Nat Biomed Eng. 2022. PMID: 35478228 Free PMC article.
-
Fluorescent hiPSC-derived MYH6-mScarlet cardiomyocytes for real-time tracking, imaging, and cardiotoxicity assays.Cell Biol Toxicol. 2023 Feb;39(1):145-163. doi: 10.1007/s10565-022-09742-0. Epub 2022 Jul 23. Cell Biol Toxicol. 2023. PMID: 35870039 Free PMC article.
-
1-deoxysphingolipids bind to COUP-TF to modulate lymphatic and cardiac cell development.Dev Cell. 2021 Nov 22;56(22):3128-3145.e15. doi: 10.1016/j.devcel.2021.10.018. Epub 2021 Nov 10. Dev Cell. 2021. PMID: 34762852 Free PMC article.
-
Electrophysiologic Characterization of Calcium Handling in Human Induced Pluripotent Stem Cell-Derived Atrial Cardiomyocytes.Stem Cell Reports. 2018 Jun 5;10(6):1867-1878. doi: 10.1016/j.stemcr.2018.04.005. Epub 2018 May 3. Stem Cell Reports. 2018. PMID: 29731429 Free PMC article.
References
-
- Birket M.J., Ribeiro M.C., Kosmidis G., Ward D., Leitoguinho A.R., van de Pol V., Dambrot C., Devalla H.D., Davis R.P., Mastroberardino P.G. Contractile defect caused by mutation in MYBPC3 revealed under conditions optimized for human PSC-cardiomyocyte function. Cell Rep. 2015;13:1–13. - PMC - PubMed
-
- Braam S.R., Tertoolen L., van de Stolpe A., Meyer T., Passier R., Mummery C.L. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. 2010;4:107–116. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

