Brain Regions Showing White Matter Loss in Huntington's Disease Are Enriched for Synaptic and Metabolic Genes
- PMID: 29174593
- PMCID: PMC5803509
- DOI: 10.1016/j.biopsych.2017.10.019
Brain Regions Showing White Matter Loss in Huntington's Disease Are Enriched for Synaptic and Metabolic Genes
Abstract
Background: The earliest white matter changes in Huntington's disease are seen before disease onset in the premanifest stage around the striatum, within the corpus callosum, and in posterior white matter tracts. While experimental evidence suggests that these changes may be related to abnormal gene transcription, we lack an understanding of the biological processes driving this regional vulnerability.
Methods: Here, we investigate the relationship between regional transcription in the healthy brain, using the Allen Institute for Brain Science transcriptome atlas, and regional white matter connectivity loss at three time points over 24 months in subjects with premanifest Huntington's disease relative to control participants. The baseline cohort included 72 premanifest Huntington's disease participants and 85 healthy control participants.
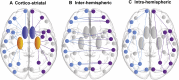
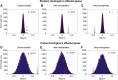
Results: We show that loss of corticostriatal, interhemispheric, and intrahemispheric white matter connections at baseline and over 24 months in premanifest Huntington's disease is associated with gene expression profiles enriched for synaptic genes and metabolic genes. Corticostriatal gene expression profiles are predominately associated with motor, parietal, and occipital regions, while interhemispheric expression profiles are associated with frontotemporal regions. We also show that genes with known abnormal transcription in human Huntington's disease and animal models are overrepresented in synaptic gene expression profiles, but not in metabolic gene expression profiles.
Conclusions: These findings suggest a dual mechanism of white matter vulnerability in Huntington's disease, in which abnormal transcription of synaptic genes and metabolic disturbance not related to transcription may drive white matter loss.
Keywords: Connectome; Genetics; Huntington's disease; Imaging; Transcription; White matter.
Copyright © 2017 Society of Biological Psychiatry. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Bates G.P., Dorsey R., Gusella J.F., Hayden M.R., Kay C., Leavitt B.R. Huntington disease. Nat Rev Dis Primers. 2015;1:15005. - PubMed
-
- Tabrizi S.J., Scahill R.I., Durr A., Roos R.A., Leavitt B.R., Jones R. Biological and clinical changes in premanifest and early stage Huntington's disease in the TRACK-HD study: The 12-month longitudinal analysis. Lancet Neurol. 2011;10:31–42. - PubMed
-
- Di Paola M., Luders E., Cherubini A., Sanchez-Castaneda C., Thompson P.M., Toga A.W. Multimodal MRI analysis of the corpus callosum reveals white matter differences in presymptomatic and early Huntington's disease. Cereb Cortex. 2012;22:2858–2866. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

