The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting
- PMID: 29174924
- PMCID: PMC5728686
- DOI: 10.1016/j.molcel.2017.10.034
The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting
Abstract
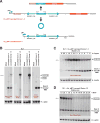
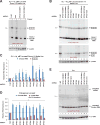
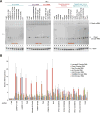
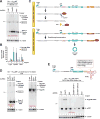
Many eukaryotic genes generate linear mRNAs and circular RNAs, but it is largely unknown how the ratio of linear to circular RNA is controlled or modulated. Using RNAi screening in Drosophila cells, we identify many core spliceosome and transcription termination factors that control the RNA outputs of reporter and endogenous genes. When spliceosome components were depleted or inhibited pharmacologically, the steady-state levels of circular RNAs increased while expression of their associated linear mRNAs concomitantly decreased. Upon inhibiting RNA polymerase II termination via depletion of the cleavage/polyadenylation machinery, circular RNA levels were similarly increased. This is because readthrough transcripts now extend into downstream genes and are subjected to backsplicing. In total, these results demonstrate that inhibition or slowing of canonical pre-mRNA processing events shifts the steady-state output of protein-coding genes toward circular RNAs. This is in part because nascent RNAs become directed into alternative pathways that lead to circular RNA production.
Keywords: Laccase2; Pladienolide B; SF3b1; backsplicing; circRNA; cleavage/polyadenylation; exon definition; pre-mRNA splicing; spliceosome; transcription termination.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Splicing: Going in circles.Nat Rev Mol Cell Biol. 2018 Jan;19(1):3. doi: 10.1038/nrm.2017.128. Epub 2017 Dec 6. Nat Rev Mol Cell Biol. 2018. PMID: 29209055 No abstract available.
-
Splicing: Going in circles.Nat Rev Genet. 2018 Feb;19(2):64-65. doi: 10.1038/nrg.2017.111. Epub 2017 Dec 18. Nat Rev Genet. 2018. PMID: 29249814 No abstract available.
References
-
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

