8-Oxoguanine DNA glycosylase 1: Beyond repair of the oxidatively modified base lesions
- PMID: 29175754
- PMCID: PMC5975208
- DOI: 10.1016/j.redox.2017.11.008
8-Oxoguanine DNA glycosylase 1: Beyond repair of the oxidatively modified base lesions
Abstract
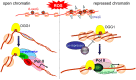
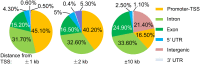
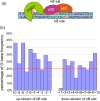
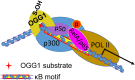
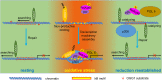
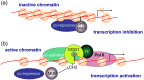
Oxidative stress and the resulting damage to genomic DNA are inevitable consequences of endogenous physiological processes, and they are amplified by cellular responses to environmental exposures. One of the most frequent reactions of reactive oxygen species with DNA is the oxidation of guanine to pre-mutagenic 8-oxo-7,8-dihydroguanine (8-oxoG). Despite the vulnerability of guanine to oxidation, vertebrate genes are primarily embedded in GC-rich genomic regions, and over 72% of the promoters of human genes belong to a class with a high GC content. In the promoter, 8-oxoG may serve as an epigenetic mark, and when complexed with the oxidatively inactivated repair enzyme 8-oxoguanine DNA glycosylase 1, provide a platform for the coordination of the initial steps of DNA repair and the assembly of the transcriptional machinery to launch the prompt and preferential expression of redox-regulated genes. Deviations/variations from this artful coordination may be the etiological links between guanine oxidation and various cellular pathologies and diseases during ageing processes.
Keywords: 8-oxoguanine; DNA methylation; Epigenetic; Gene expression regulation; OGG1.
Copyright © 2017 The Authors. Published by Elsevier B.V. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

