Deciphering cyanobacterial phenotypes for fast photoautotrophic growth via isotopically nonstationary metabolic flux analysis
- PMID: 29177008
- PMCID: PMC5691832
- DOI: 10.1186/s13068-017-0958-y
Deciphering cyanobacterial phenotypes for fast photoautotrophic growth via isotopically nonstationary metabolic flux analysis
Abstract
Background: Synechococcus elongatus UTEX 2973 is the fastest growing cyanobacterium characterized to date. Its genome was found to be 99.8% identical to S. elongatus 7942 yet it grows twice as fast. Current genome-to-phenome mapping is still poorly performed for non-model organisms. Even for species with identical genomes, cell phenotypes can be strikingly different. To understand Synechococcus 2973's fast-growth phenotype and its metabolic features advantageous to photo-biorefineries, 13C isotopically nonstationary metabolic flux analysis (INST-MFA), biomass compositional analysis, gene knockouts, and metabolite profiling were performed on both strains under various growth conditions.
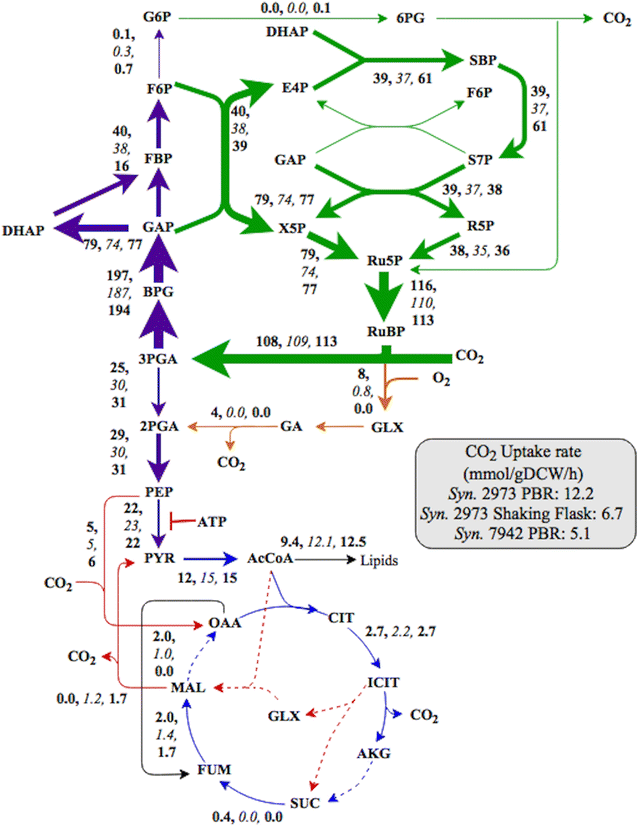
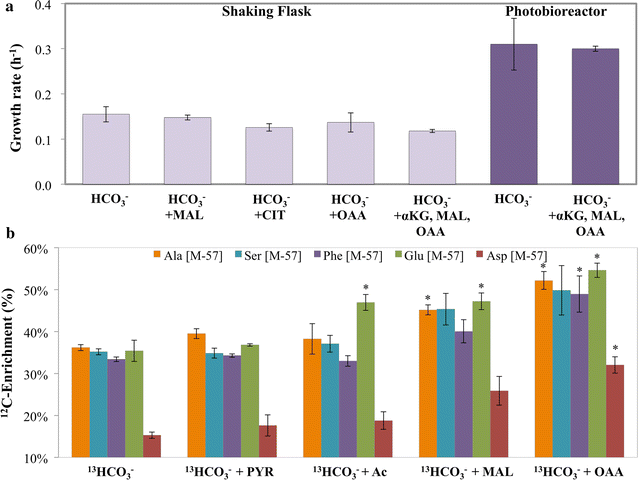
Results: The Synechococcus 2973 flux maps show substantial carbon flow through the Calvin cycle, glycolysis, photorespiration and pyruvate kinase, but minimal flux through the malic enzyme and oxidative pentose phosphate pathways under high light/CO2 conditions. During fast growth, its pool sizes of key metabolites in central pathways were lower than suboptimal growth. Synechococcus 2973 demonstrated similar flux ratios to Synechococcus 7942 (under fast growth conditions), but exhibited greater carbon assimilation, higher NADPH concentrations, higher energy charge (relative ATP ratio over ADP and AMP), less accumulation of glycogen, and potentially metabolite channeling. Furthermore, Synechococcus 2973 has very limited flux through the TCA pathway with small pool sizes of acetyl-CoA/TCA intermediates under all growth conditions.
Conclusions: This study employed flux analysis to investigate phenotypic heterogeneity among two cyanobacterial strains with near-identical genome background. The flux/metabolite profiling, biomass composition analysis, and genetic modification results elucidate a highly effective metabolic topology for CO2 assimilatory and biosynthesis in Synechococcus 2973. Comparisons across multiple Synechococcus strains indicate faster metabolism is also driven by proportional increases in both photosynthesis and key central pathway fluxes. Moreover, the flux distribution in Synechococcus 2973 supports the use of its strong sugar phosphate pathways for optimal bio-productions. The integrated methodologies in this study can be applied for characterizing non-model microbial metabolism.
Keywords: 13C labeling experiments; Channeling; Energy charge; Glycogen; Metabolites; Photobioreactor.
Figures



References
-
- Microalgae Sayre R. The potential for carbon capture. Bioscience. 2010;60:722–727. doi: 10.1525/bio.2010.60.9.9. - DOI
-
- Xiong W, Morgan JA, Ungerer J, Wang B, Maness PC, Yu J. The plasticity of cyanobacterial metabolism supports direct CO2 conversion to ethylene. Nat Plants. 2015;1:15053. doi: 10.1038/nplants.2015.53. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

