ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads
- PMID: 29177089
- PMCID: PMC5695208
- DOI: 10.1099/mgen.0.000131
ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads
Abstract
Antimicrobial resistance (AMR) is one of the major threats to human and animal health worldwide, yet few high-throughput tools exist to analyse and predict the resistance of a bacterial isolate from sequencing data. Here we present a new tool, ARIBA, that identifies AMR-associated genes and single nucleotide polymorphisms directly from short reads, and generates detailed and customizable output. The accuracy and advantages of ARIBA over other tools are demonstrated on three datasets from Gram-positive and Gram-negative bacteria, with ARIBA outperforming existing methods.
Keywords: antimicrobial resistance; bacteria; genotyping; sequence typing; whole genome sequencing.
Figures

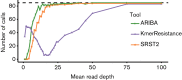


References
-
- O'Neill J. Tackling drug-resistant infections globally: Final report and recommendations. The review on antimicrobial resistance. London: HM Government and the Wellcome Trust; 2016. https://amr-review.org/sites/default/files/160525_Final%20paper_with%20c...
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

