Statistical description of the denatured structure of a single protein, staphylococcal nuclease, by FRET analysis
- PMID: 29178080
- PMCID: PMC5899696
- DOI: 10.1007/s12551-017-0334-y
Statistical description of the denatured structure of a single protein, staphylococcal nuclease, by FRET analysis
Abstract
Structural characterization of fully unfolded proteins is essential for understanding not only protein-folding mechanisms, but also the structures of intrinsically disordered proteins. Because an unfolded protein can assume all possible conformations, statistical descriptions of its structure are most appropriate. For this purpose, we applied Förster resonance energy transfer (FRET) analysis to fully unfolded staphylococcal nuclease. Artificial amino acids labeled with a FRET donor or acceptor were introduced by an amber codon and a four-base codon respectively. Eight double-labeled proteins were prepared, purified, and subjected to FRET analysis in 6 M urea. The observed behavior could be explained by a power law, R = αN0.44, where R, and N are the distance and the number of residues between donor and acceptor, and α is a coefficient. The index was smaller than the value expected for an excluded-volume random coil, 0.588, indicating that the fully unfolded proteins were more compact than polypeptides in good solvent. The FRET efficiency in the native state did not necessarily correlate to the distance obtained from crystal structure, suggesting that other factors such as the orientation factor made a substantial contribution to FRET.
Keywords: Denatured structure; FRET; Protein; Random coil; Staphylococcal nuclease.
Conflict of interest statement
Conflict of interest
All authors declare that they have no conflict of interest
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Figures

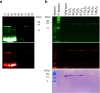

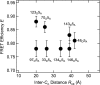

References
-
- De Gennes PG (1979) Scaling concepts in polymer physics. Cornell University Press, Ithaca NY
-
- Dill KA. Folding proteins: finding a needle in a haystack. Curr Opin Struct Biol. 1993;3:99–103. doi: 10.1016/0959-440X(93)90208-3. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

