Cohort-specific imputation of gene expression improves prediction of warfarin dose for African Americans
- PMID: 29178968
- PMCID: PMC5702158
- DOI: 10.1186/s13073-017-0495-0
Cohort-specific imputation of gene expression improves prediction of warfarin dose for African Americans
Abstract
Background: Genome-wide association studies are useful for discovering genotype-phenotype associations but are limited because they require large cohorts to identify a signal, which can be population-specific. Mapping genetic variation to genes improves power and allows the effects of both protein-coding variation as well as variation in expression to be combined into "gene level" effects.
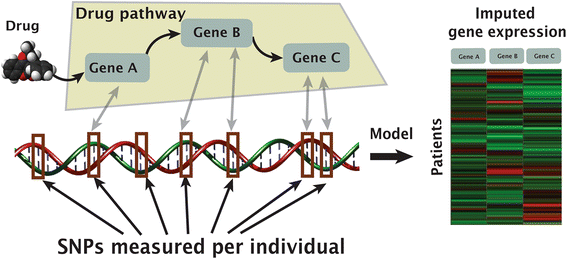
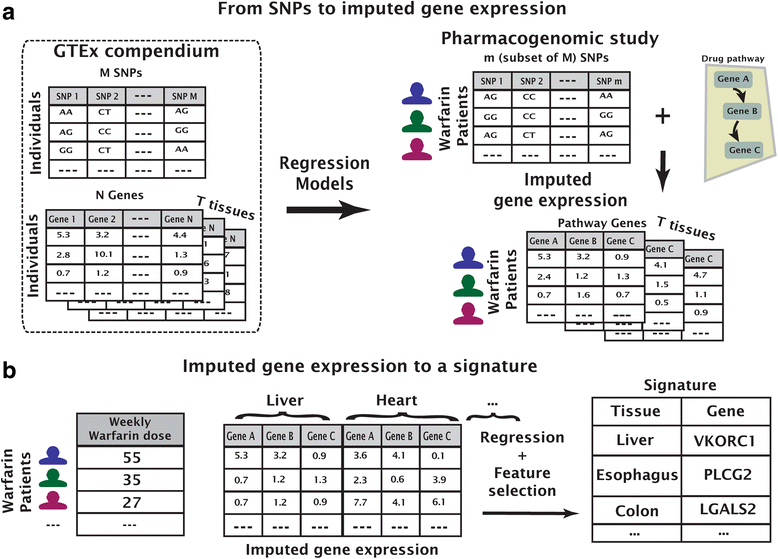
Methods: Previous work has shown that warfarin dose can be predicted using information from genetic variation that affects protein-coding regions. Here, we introduce a method that improves dose prediction by integrating tissue-specific gene expression. In particular, we use drug pathways and expression quantitative trait loci knowledge to impute gene expression-on the assumption that differential expression of key pathway genes may impact dose requirement. We focus on 116 genes from the pharmacokinetic and pharmacodynamic pathways of warfarin within training and validation sets comprising both European and African-descent individuals.
Results: We build gene-tissue signatures associated with warfarin dose in a cohort-specific manner and identify a signature of 11 gene-tissue pairs that significantly augments the International Warfarin Pharmacogenetics Consortium dosage-prediction algorithm in both populations.
Conclusions: Our results demonstrate that imputed expression can improve dose prediction and bridge population-specific compositions. MATLAB code is available at https://github.com/assafgo/warfarin-cohort.
Keywords: African Americans; International Warfarin Pharmacogenetics Consortium; Pharmacogenomics; Warfarin dose.
Conflict of interest statement
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
MeSH terms
Substances
Grants and funding
- R24 GM061374/GM/NIGMS NIH HHS/United States
- 521-2014-3370/Vetenskapsrådet
- R01 HG008150/HG/NHGRI NIH HHS/United States
- U01HG007436/National Human Genome Research Institute
- R01 LM005652/LM/NLM NIH HHS/United States
- LM05652/U.S. National Library of Medicine
- RG/14/5/30893/British Heart Foundation/United Kingdom
- 20120557/Hjärt-Lungfonden
- 20140291/Hjärt-Lungfonden
- U01 GM061374/GM/NIGMS NIH HHS/United States
- R01 GM102365/GM/NIGMS NIH HHS/United States
- 521-2011-2440/Vetenskapsrådet
- U01 HG007436/HG/NHGRI NIH HHS/United States
- GM061374/National Institute of General Medical Sciences
- R01MH101814/National Institute of Mental Health
- R01 MH101814/MH/NIMH NIH HHS/United States
- U01HG00908001/National Human Genome Research Institute
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

