When DNA Topology Turns Deadly - RNA Polymerases Dig in Their R-Loops to Stand Their Ground: New Positive and Negative (Super)Twists in the Replication-Transcription Conflict
- PMID: 29179918
- PMCID: PMC5967978
- DOI: 10.1016/j.tig.2017.10.007
When DNA Topology Turns Deadly - RNA Polymerases Dig in Their R-Loops to Stand Their Ground: New Positive and Negative (Super)Twists in the Replication-Transcription Conflict
Abstract
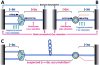
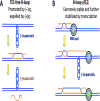
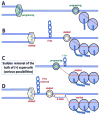
Head-on replication-transcription conflict is especially bitter in bacterial chromosomes, explaining why actively transcribed genes are always co-oriented with replication. The mechanism of this conflict remains unclear, besides the anticipated accumulation of positive supercoils between head-on-conflicting polymerases. Unexpectedly, experiments in bacterial and human cells reveal that head-on replication-transcription conflict induces R-loops, indicating hypernegative supercoiling [(-)sc] in the region - precisely the opposite of that assumed. Further, as a result of these R-loops, both replication and transcription in the affected region permanently stall, so the failure of R-loop removal in RNase H-deficient bacteria becomes lethal. How hyper(-)sc emerges in the middle of a positively supercoiled chromosomal domain is a mystery that requires rethinking of topoisomerase action around polymerases.
Keywords: R-loops; RNase H; replication–transcription conflicts; supercoiling; topoisomerases.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures

References
-
- Brewer BJ. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988;53:679–686. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

