The ABC of Biofilm Drug Tolerance: the MerR-Like Regulator BrlR Is an Activator of ABC Transport Systems, with PA1874-77 Contributing to the Tolerance of Pseudomonas aeruginosa Biofilms to Tobramycin
- PMID: 29180529
- PMCID: PMC5786766
- DOI: 10.1128/AAC.01981-17
The ABC of Biofilm Drug Tolerance: the MerR-Like Regulator BrlR Is an Activator of ABC Transport Systems, with PA1874-77 Contributing to the Tolerance of Pseudomonas aeruginosa Biofilms to Tobramycin
Abstract
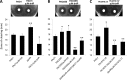
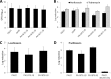
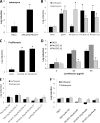
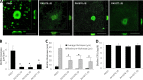
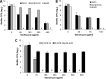
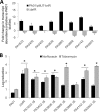
A hallmark of biofilms is their tolerance to killing by antimicrobial agents. In Pseudomonas aeruginosa, biofilm drug tolerance requires the c-di-GMP-responsive MerR transcriptional regulator BrlR. However, the mechanism by which BrlR mediates biofilm drug tolerance has not been elucidated. Here, we demonstrate that BrlR activates the expression of at least 7 ABC transport systems, including the PA1874-PA1875-PA1876-PA1877 (PA1874-77) operon, with chromatin immunoprecipitation and DNA binding assays confirming BrlR binding to the promoter region of PA1874-77. Insertional inactivation of the 7 ABC transport systems rendered P. aeruginosa PAO1 biofilms susceptible to tobramycin or norfloxacin. Susceptibility was linked to drug accumulation, with BrlR contributing to norfloxacin accumulation in a manner dependent on multidrug efflux pumps and the PA1874-77 ABC transport system. Inactivation of the respective ABC transport system, furthermore, eliminated the recalcitrance of biofilms to killing by tobramycin but not norfloxacin, indicating that drug accumulation is not linked to biofilm drug tolerance. Our findings indicate for the first time that BrlR, a MerR-type transcriptional activator, activates genes encoding several ABC transport systems, in addition to multiple multidrug efflux pump genes. Moreover, our data confirm a BrlR target contributing to drug tolerance, likely countering the prevailing dogma that biofilm tolerance arises from a multiplicity of factors.
Keywords: ABC transport system; ABC transporter; MBC biofilm assay; MexAB-OprM; PA4142-44; PA5503-05; biofilm MBC; biofilm drug tolerance; drug accumulation; kdpFABC; opuC; phosphotransfer.
Copyright © 2018 American Society for Microbiology.
Figures

Similar articles
-
FleQ finetunes the expression of a subset of BrlR-activated genes to enable antibiotic tolerance by Pseudomonas aeruginosa biofilms.J Bacteriol. 2025 May 22;207(5):e0050324. doi: 10.1128/jb.00503-24. Epub 2025 Apr 30. J Bacteriol. 2025. PMID: 40304498 Free PMC article.
-
The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug efflux pumps in Pseudomonas aeruginosa biofilms.J Bacteriol. 2013 Aug;195(15):3352-63. doi: 10.1128/JB.00318-13. Epub 2013 May 17. J Bacteriol. 2013. PMID: 23687276 Free PMC article.
-
The MerR-like regulator BrlR impairs Pseudomonas aeruginosa biofilm tolerance to colistin by repressing PhoPQ.J Bacteriol. 2013 Oct;195(20):4678-88. doi: 10.1128/JB.00834-13. Epub 2013 Aug 9. J Bacteriol. 2013. PMID: 23935054 Free PMC article.
-
The PA3177 Gene Encodes an Active Diguanylate Cyclase That Contributes to Biofilm Antimicrobial Tolerance but Not Biofilm Formation by Pseudomonas aeruginosa.Antimicrob Agents Chemother. 2018 Sep 24;62(10):e01049-18. doi: 10.1128/AAC.01049-18. Print 2018 Oct. Antimicrob Agents Chemother. 2018. PMID: 30082282 Free PMC article.
-
Efflux systems as a target for anti-biofilm nanoparticles: perspectives on emerging applications.Expert Opin Ther Targets. 2023 Jul-Dec;27(10):953-963. doi: 10.1080/14728222.2023.2263910. Epub 2023 Oct 30. Expert Opin Ther Targets. 2023. PMID: 37788168 Review.
Cited by
-
Role of Efflux Pumps on Antimicrobial Resistance in Pseudomonas aeruginosa.Int J Mol Sci. 2022 Dec 13;23(24):15779. doi: 10.3390/ijms232415779. Int J Mol Sci. 2022. PMID: 36555423 Free PMC article. Review.
-
Controlling Biofilm Development Through Cyclic di-GMP Signaling.Adv Exp Med Biol. 2022;1386:69-94. doi: 10.1007/978-3-031-08491-1_3. Adv Exp Med Biol. 2022. PMID: 36258069 Free PMC article. Review.
-
Overexpression of BIT33_RS14560 Enhances the Biofilm Formation and Virulence of Acinetobacter baumannii.Front Microbiol. 2022 Apr 25;13:867770. doi: 10.3389/fmicb.2022.867770. eCollection 2022. Front Microbiol. 2022. PMID: 35547150 Free PMC article.
-
Flip the switch: the role of FleQ in modulating the transition between the free-living and sessile mode of growth in Pseudomonas aeruginosa.J Bacteriol. 2024 Mar 21;206(3):e0036523. doi: 10.1128/jb.00365-23. Epub 2024 Mar 4. J Bacteriol. 2024. PMID: 38436566 Free PMC article. Review.
-
Bacterial MerR family transcription regulators: activationby distortion.Acta Biochim Biophys Sin (Shanghai). 2022 Jan 25;54(1):25-36. doi: 10.3724/abbs.2021003. Acta Biochim Biophys Sin (Shanghai). 2022. PMID: 35130613 Free PMC article. Review.
References
-
- Hall-Stoodley L, Hu FZ, Gieseke A, Nistico L, Nguyen D, Hayes J, Forbes M, Greenberg DP, Dice B, Burrows A, Wackym PA, Stoodley P, Post JC, Ehrlich GD, Kerschner JE. 2006. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296:202–211. doi:10.1001/jama.296.2.202. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

