Radiotherapy and CTLA-4 Blockade Shape the TCR Repertoire of Tumor-Infiltrating T Cells
- PMID: 29180535
- PMCID: PMC6020019
- DOI: 10.1158/2326-6066.CIR-17-0134
Radiotherapy and CTLA-4 Blockade Shape the TCR Repertoire of Tumor-Infiltrating T Cells
Abstract
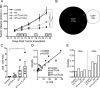
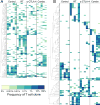
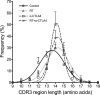
Immune checkpoint inhibitors activate T cells to reject tumors. Unique tumor mutations are key T-cell targets, but a comprehensive understanding of the nature of a successful antitumor T-cell response is lacking. To investigate the T-cell receptor (TCR) repertoire associated with treatment success versus failure, we used a well-characterized mouse carcinoma that is rejected by CD8 T cells in mice treated with radiotherapy (RT) and anti-CTLA-4 in combination, but not as monotherapy, and comprehensively analyzed tumor-infiltrating lymphocytes (TILs) by high-throughput sequencing of the TCRΒ CDR3 region. The combined treatment increased TIL density and CD8/CD4 ratio. Assessment of the frequency of T-cell clones indicated that anti-CTLA-4 resulted in fewer clones and a more oligoclonal repertoire compared with untreated tumors. In contrast, RT increased the CD8/CD4 ratio and broadened the TCR repertoire, and when used in combination with anti-CTLA-4, these selected T-cell clones proliferated. Hierarchical clustering of CDR3 sequences showed a treatment-specific clustering of TCRs that were shared by different mice. Abundant clonotypes were commonly shared between animals and yet treatment-specific. Analysis of amino-acid sequence similarities revealed a significant increase in the number and richness of dominant CDR3 motifs in tumors treated with RT + anti-CTLA-4 compared with control. The repertoire of TCRs reactive with a single tumor antigen recognized by CD8+ T cells was heterogeneous but highly clonal, irrespective of treatment. Overall, data support a model whereby a diverse TCR repertoire is required to achieve tumor rejection and may underlie the synergy between RT and CTLA-4 blockade. Cancer Immunol Res; 6(2); 139-50. ©2017 AACR.
©2017 American Association for Cancer Research.
Conflict of interest statement
Figures
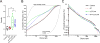


References
-
- Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

