Mycosphaerellaceae - Chaos or clarity?
- PMID: 29180830
- PMCID: PMC5693839
- DOI: 10.1016/j.simyco.2017.09.003
Mycosphaerellaceae - Chaos or clarity?
Abstract
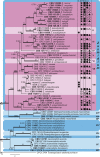
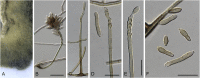
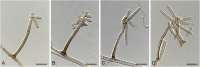
The Mycosphaerellaceae represent thousands of fungal species that are associated with diseases on a wide range of plant hosts. Understanding and stabilising the taxonomy of genera and species of Mycosphaerellaceae is therefore of the utmost importance given their impact on agriculture, horticulture and forestry. Based on previous molecular studies, several phylogenetic and morphologically distinct genera within the Mycosphaerellaceae have been delimited. In this study a multigene phylogenetic analysis (LSU, ITS and rpb2) was performed based on 415 isolates representing 297 taxa and incorporating ex-type strains where available. The main aim of this study was to resolve the phylogenetic relationships among the genera currently recognised within the family, and to clarify the position of the cercosporoid fungi among them. Based on these results many well-known genera are shown to be paraphyletic, with several synapomorphic characters that have evolved more than once within the family. As a consequence, several old generic names including Cercosporidium, Fulvia, Mycovellosiella, Phaeoramularia and Raghnildiana are resurrected, and 32 additional genera are described as new. Based on phylogenetic data 120 genera are now accepted within the family, but many currently accepted cercosporoid genera still remain unresolved pending fresh collections and DNA data. The present study provides a phylogenetic framework for future taxonomic work within the Mycosphaerellaceae.
Keywords: Adelopus gaeumannii T. Rohde; Amycosphaerella keniensis (Crous & T.A. Cout.) Videira & Crous; Australosphaerella Videira & Crous; Australosphaerella nootherensis (Carnegie) Videira & Crous; Biharia vangueriae Thirum. & Mishra; Brunswickiella Videira & Crous; Brunswickiella parsonsiae (Crous & Summerell) Videira & Crous; Catenulocercospora C. Nakash., Videira & Crous; Catenulocercospora fusimaculans (G.F. Atk.) C. Nakash., Videira & Crous; Cercoramularia Videira, H.D. Shin, C. Nakash. & Crous; Cercoramularia koreana Videira, H.D. Shin, C. Nakash. & Crous; Cercospora brachycarpa Syd.; Cercospora cajani Henn.; Cercospora desmodii Ellis & Kellerm.; Cercospora ferruginea Fuckel; Cercospora gnaphaliacea Cooke; Cercospora gomphrenicola Speg.; Cercospora henningsii Allesch.; Cercospora mangiferae Koord.; Cercospora microsora Sacc.; Cercospora rosicola Pass.; Cercospora smilacis Thüm.; Cercospora tiliae Peck; Cercosporidium californicum (S.T. Koike & Crous) Videira & Crous; Cercosporidium helleri Earle; Chuppomyces Videira & Crous; Chuppomyces handelii (Bubák) U. Braun, C. Nakash., Videira & Crous; Cladosporium bacilligerum Mont. & Fr.; Cladosporium chaetomium Cooke; Cladosporium fulvum Cooke; Cladosporium lonicericola Yong H. He & Z.Y. Zhang; Cladosporium personatum Berk. & M.A. Curtis; Clarohilum Videira & Crous; Clarohilum henningsii (Allesch.) Videira & Crous; Clasterosporium degenerans Syd. & P. Syd.; Clypeosphaerella calotropidis (Ellis & Everh.) Videira & Crous; Collarispora Videira & Crous; Collarispora valgourgensis (Crous) Videira & Crous; Coremiopassalora U. Braun, C. Nakash., Videira & Crous; Coremiopassalora eucalypti (Crous & Alfenas) U. Braun, C. Nakash., Videira & Crous; Coremiopassalora leptophlebae (Crous et al.) U. Braun, C. Nakash., Videira & Crous; Coryneum vitiphyllum Speschnew; Cryptosporium acicola Thüm.; Deightonomyces Videira & Crous; Deightonomyces daleae (Ellis & Kellerm.) Videira & Crous; Devonomyces Videira & Crous; Devonomyces endophyticus (Crous & H. Sm. Ter) Videira & Crous; Distocercosporaster Videira, H.D. Shin, C. Nakash. & Crous; Distocercosporaster dioscoreae (Ellis & G. Martin) Videira, H.D. Shin, C. Nakash. & Crous; Distomycovellosiella U. Braun, C. Nakash., Videira & Crous; Distomycovellosiella brachycarpa (Syd.) U. Braun, C. Nakash., Videira & Crous; Exopassalora Videira & Crous; Exopassalora zambiae (Crous & T.A. Cout.) Videira & Crous; Exosporium livistonicola U. Braun, Videira & Crous for Distocercospora livistonae U. Braun & C.F. Hill; Exutisphaerella Videira & Crous; Exutisphaerella laricina (R. Hartig) Videira & Crous; Fusoidiella anethi (Pers.) Videira & Crous; Graminopassalora U. Braun, C. Nakash., Videira & Crous; Graminopassalora graminis (Fuckel) U. Braun, C. Nakash., Videira & Crous; Helicoma fasciculatum Berk. & M.A. Curtis.; Hyalocercosporidium Videira & Crous; Hyalocercosporidium desmodii Videira & Crous; Hyalozasmidium U. Braun, C. Nakash., Videira & Crous; Hyalozasmidium aerohyalinosporum (Crous & Summerell) Videira & Crous; Hyalozasmidium sideroxyli U. Braun, C. Nakash., Videira & Crous; Isariopsis griseola Sacc.; Madagascaromyces U. Braun, C. Nakash., Videira & Crous; Madagascaromyces intermedius (Crous & M.J. Wingf.) Videira & Crous; Micronematomyces U. Braun, C. Nakash., Videira & Crous; Micronematomyces caribensis (Crous & Den Breeÿen) U. Braun, C. Nakash., Videira & Crous; Micronematomyces chromolaenae (Crous & Den Breeÿen) U. Braun, C. Nakash., Videira & Crous; Multi-gene phylogeny; Mycosphaerella; Neoceratosperma haldinae U. Braun, C. Nakash., Videira & Crous; Neoceratosperma legnephoricola U. Braun, C. Nakash., Videira & Crous; Neocercosporidium Videira & Crous; Neocercosporidium smilacis (Thüm.) U. Braun, C. Nakash., Videira & Crous; Neophloeospora Videira & Crous; Neophloeospora maculans (Bérenger) Videira & Crous; Nothopassalora U. Braun, C. Nakash., Videira & Crous; Nothopassalora personata (Berk. & M.A. Curtis) U. Braun, C. Nakash., Videira & Crous; Nothopericoniella Videira & Crous; Nothopericoniella perseae-macranthae (Hosag. & U. Braun) Videira & Crous; Nothophaeocryptopus Videira, C. Nakash., U. Braun, Crous; Nothophaeocryptopus gaeumannii (T. Rohde) Videira, C. Nakash., U. Braun, Crous; Pachyramichloridium Videira & Crous; Pachyramichloridium pini (de Hoog & Rahman) U. Braun, C. Nakash., Videira & Crous; Paracercosporidium Videira & Crous; Paracercosporidium microsorum (Sacc.) U. Braun, C. Nakash., Videira & Crous; Paracercosporidium tiliae (Peck) U. Braun, C. Nakash., Videira & Crous; Paramycosphaerella wachendorfiae (Crous) Videira & Crous; Paramycovellosiella Videira, H.D. Shin & Crous; Paramycovellosiella passaloroides (G. Winter) Videira, H.D. Shin & Crous; Parapallidocercospora Videira, Crous, U. Braun, C. Nakash.; Parapallidocercospora colombiensis (Crous et al.) Videira & Crous; Parapallidocercospora thailandica (Crous et al.) Videira & Crous; Phaeocercospora juniperina (Georgescu & Badea) U. Braun, C. Nakash., Videira & Crous; Plant pathogen; Pleopassalora Videira & Crous; Pleopassalora perplexa (Beilharz et al.) Videira & Crous; Pleuropassalora U. Braun, C. Nakash., Videira & Crous; Pleuropassalora armatae (Crous & A.R. Wood) U. Braun, C. Nakash., Videira & Crous; Pluripassalora Videira & Crous; Pluripassalora bougainvilleae (Munt.-Cvetk.) U. Braun, C. Nakash., Videira & Crous; Pseudocercospora convoluta (Crous & Den Breeÿen) U. Braun, C. Nakash., Videira & Crous; Pseudocercospora nodosa (Constant.) U. Braun, C. Nakash., Videira & Crous; Pseudocercospora platanigena Videira & Crous for Stigmella platani Fuckel, non Pseudocercospora platani (J.M. Yen) J.M. Yen 1979; Pseudocercospora zambiensis (Deighton) Crous & U. Braun; Pseudopericoniella Videira & Crous; Pseudopericoniella levispora (Arzanlou, W. Gams & Crous) Videira & Crous; Pseudophaeophleospora U. Braun, C. Nakash., Videira & Crous; Pseudophaeophleospora atkinsonii (Syd.) U. Braun, C. Nakash., Videira & Crous; Pseudophaeophleospora stonei (Crous) U. Braun, C. Nakash., Videira & Crous; Pseudozasmidium Videira & Crous; Pseudozasmidium eucalypti (Crous & Summerell) Videira & Crous; Pseudozasmidium nabiacense (Crous & Carnegie) Videira & Crous; Pseudozasmidium parkii (Crous & Alfenas) Videira & Crous; Pseudozasmidium vietnamense (Barber & T.I. Burgess) Videira & Crous; Ragnhildiana ampelopsidis (Peck) U. Braun, C. Nakash., Videira & Crous; Ragnhildiana diffusa (Heald & F.A. Wolf) Videira & Crous; Ragnhildiana ferruginea (Fuckel) U. Braun, C. Nakash., Videira & Crous; Ragnhildiana gnaphaliaceae (Cooke) Videira, H.D. Shin, C. Nakash. & Crous; Ragnhildiana perfoliati (Ellis & Everh.) U. Braun, C. Nakash., Videira & Crous; Ragnhildiana pseudotithoniae (Crous & Cheew.) U. Braun, C. Nakash., Videira & Crous; Ramulispora sorghiphila U. Braun, C. Nakash., Videira & Crous; Rhachisphaerella Videira & Crous; Rhachisphaerella mozambica (Arzanlou & Crous) Videira & Crous; Rosisphaerella Videira & Crous; Rosisphaerella rosicola (Pass.) U. Braun, C. Nakash., Videira & Crous; Scolicotrichum roumeguerei Briosi & Cavara; Septoria martiniana Sacc; Sphaerella araneosa Rehm; Sphaerella laricina R. Hartig; Stictosepta cupularis Petr.; Stigmella platani Fuckel; Sultanimyces Videira & Crous; Sultanimyces vitiphyllus (Speschnew) Videira & Crous; Tapeinosporium viride Bonord; Taxonomy; Utrechtiana roumeguerei (Cavara) Videira & Crous; Virosphaerella Videira & Crous; Virosphaerella irregularis (Cheew. et al.) Videira & Crous; Virosphaerella pseudomarksii (Cheew. et al.) Videira & Crous; Xenosonderhenioides Videira & Crous; Xenosonderhenioides indonesiana C. Nakash., Videira & Crous; Zasmidium arcuatum (Arzanlou et al.) Videira & Crous; Zasmidium biverticillatum (Arzanlou & Crous) Videira & Crous; Zasmidium cerophilum (Tubaki) U. Braun, C. Nakash., Videira & Crous; Zasmidium daviesiae (Cooke & Massee) U. Braun, C. Nakash., Videira & Crous; Zasmidium elaeocarpi U. Braun, C. Nakash., Videira & Crous; Zasmidium eucalypticola U. Braun, C. Nakash., Videira & Crous; Zasmidium grevilleae U. Braun, C. Nakash., Videira & Crous; Zasmidium gupoyu (R. Kirschner) U. Braun, C. Nakash., Videira & Crous; Zasmidium hakeae U. Braun, C. Nakash., Videira & Crous; Zasmidium iteae (R. Kirschner) U. Braun, C. Nakash., Videira & Crous; Zasmidium musae-banksii Videira & Crous for Ramichloridium australiense Arzanlou & Crous, non Zasmidium australiense (J.L. Mulder) U. Braun & Crous 2013; Zasmidium musigenum Videira & Crous for Veronaea musae Stahel ex M.B. Ellis, non Zasmidium musae (Arzanlou & Crous) Crous & U. Braun 2010; Zasmidium proteacearum (D.E. Shaw & Alcorn) U. Braun, C. Nakash. & Crous; Zasmidium pseudotsugae (V.A.M. Mill. & Bonar) Videira & Crous; Zasmidium pseudovespa (Carnegie) U. Braun, C. Nakash., Videira & Crous; Zasmidium schini U. Braun, C. Nakash., Videira & Crous; Zasmidium strelitziae (Arzanlou et al.) Videira & Crous; Zasmidium tsugae (Dearn.) Videira & Crous; Zasmidium velutinum (G. Winter) Videira & Crous.
Figures















































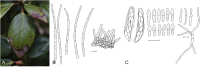



















Similar articles
-
Phylogenetic lineages in Pseudocercospora.Stud Mycol. 2013 Jun 30;75(1):37-114. doi: 10.3114/sim0005. Stud Mycol. 2013. PMID: 24014898 Free PMC article.
-
Genera of phytopathogenic fungi: GOPHY 4.Stud Mycol. 2022 Jul;101:417-564. doi: 10.3114/sim.2022.101.06. Epub 2022 Jun 2. Stud Mycol. 2022. PMID: 36059898 Free PMC article.
-
Fusarium: more than a node or a foot-shaped basal cell.Stud Mycol. 2021 Aug 17;98:100116. doi: 10.1016/j.simyco.2021.100116. eCollection 2021 Apr. Stud Mycol. 2021. PMID: 34466168 Free PMC article.
-
Redefining the megagenus Erica L. (Ericaceae): the contributions of E. G. H. Oliver and I. M. Oliver (née Nitzsche) to taxonomy and nomenclature.PhytoKeys. 2024 Jul 5;244:39-55. doi: 10.3897/phytokeys.244.121705. eCollection 2024. PhytoKeys. 2024. PMID: 39006939 Free PMC article. Review.
-
Dr. John Cooke on the Various Species of Palsy.Med Chir Rev J Med Sci Anal Ser. 1821 Mar 1;1(4):725-743. Med Chir Rev J Med Sci Anal Ser. 1821. PMID: 29917411 Free PMC article. Review. No abstract available.
Cited by
-
Taxonomy and phylogeny of cercosporoid ascomycetes on Diospyros spp. with special emphasis on Pseudocercospora spp.Fungal Syst Evol. 2020 Dec;6:95-127. doi: 10.3114/fuse.2020.06.06. Epub 2020 Apr 3. Fungal Syst Evol. 2020. PMID: 32904397 Free PMC article.
-
Fungal Planet description sheets: 868-950.Persoonia. 2019 Jun;42:291-473. doi: 10.3767/persoonia.2019.42.11. Epub 2019 Jul 19. Persoonia. 2019. PMID: 31551622 Free PMC article.
-
101 Dothideomycetes genomes: A test case for predicting lifestyles and emergence of pathogens.Stud Mycol. 2020 Feb 1;96:141-153. doi: 10.1016/j.simyco.2020.01.003. eCollection 2020 Jun. Stud Mycol. 2020. PMID: 32206138 Free PMC article.
-
Fungal Planet description sheets: 951-1041.Persoonia. 2019;43:223-425. doi: 10.3767/persoonia.2019.43.06. Epub 2019 Dec 18. Persoonia. 2019. PMID: 32214501 Free PMC article.
-
Foliar pathogens of eucalypts.Stud Mycol. 2019 Aug 8;94:125-298. doi: 10.1016/j.simyco.2019.08.001. eCollection 2019 Sep. Stud Mycol. 2019. PMID: 31636729 Free PMC article.
References
-
- Aly A.H., Debbab A., Proksch P. Fungal endophytes: unique plant inhabitants with great promises. Applied Microbiology Biotechnology. 2012;90:1829–1845. - PubMed
-
- Aptroot A. Mycosphaerella and its anamorphs 2. Conspectus of Mycosphaerella. CBS Biodiversity series. 2006;5:1–231.
-
- Arnold R.H. The Nectriella element of “Hyalodothis”. Mycologia. 1967;59:246–254.
-
- Arnold R.H. Melanodothis caricis, n. gen., n. sp. and 'Hyalodothis? caricis'. Canadian Journal of Botany. 1971;49(12):2187–2196.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
