Monitoring transcription initiation activities in rat and dog
- PMID: 29182598
- PMCID: PMC5704677
- DOI: 10.1038/sdata.2017.173
Monitoring transcription initiation activities in rat and dog
Abstract
The promoter landscape of several non-human model organisms is far from complete. As a part of FANTOM5 data collection, we generated 13 profiles of transcription initiation activities in dog and rat aortic smooth muscle cells, mesenchymal stem cells and hepatocytes by employing CAGE (Cap Analysis of Gene Expression) technology combined with single molecule sequencing. Our analyses show that the CAGE profiles recapitulate known transcription start sites (TSSs) consistently, in addition to uncover novel TSSs. Our dataset can be thus used with high confidence to support gene annotation in dog and rat species. We identified 28,497 and 23,147 CAGE peaks, or promoter regions, for rat and dog respectively, and associated them to known genes. This approach could be seen as a standard method for improvement of existing gene models, as well as discovery of novel genes. Given that the FANTOM5 data collection includes dog and rat matched cell types in human and mouse as well, this data would also be useful for cross-species studies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
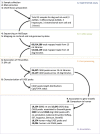


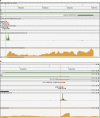
References
Data Citations
References
-
- Fang Z. & Cui X. Design and validation issues in RNA-seq experiments. Brief Bioinform 12, 280–287 (2011). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

