Vaccination with Recombinant Cryptococcus Proteins in Glucan Particles Protects Mice against Cryptococcosis in a Manner Dependent upon Mouse Strain and Cryptococcal Species
- PMID: 29184017
- PMCID: PMC5705919
- DOI: 10.1128/mBio.01872-17
Vaccination with Recombinant Cryptococcus Proteins in Glucan Particles Protects Mice against Cryptococcosis in a Manner Dependent upon Mouse Strain and Cryptococcal Species
Abstract
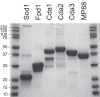
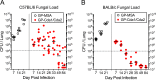
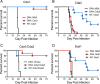
Development of a vaccine to protect against cryptococcosis is a priority given the enormous global burden of disease in at-risk individuals. Using glucan particles (GPs) as a delivery system, we previously demonstrated that mice vaccinated with crude Cryptococcus-derived alkaline extracts were protected against lethal challenge with Cryptococcus neoformans and Cryptococcus gattii The goal of the present study was to identify protective protein antigens that could be used in a subunit vaccine. Using biased and unbiased approaches, six candidate antigens (Cda1, Cda2, Cda3, Fpd1, MP88, and Sod1) were selected, recombinantly expressed in Escherichia coli, purified, and loaded into GPs. Three mouse strains (C57BL/6, BALB/c, and DR4) were then vaccinated with the antigen-laden GPs, following which they received a pulmonary challenge with virulent C. neoformans and C. gattii strains. Four candidate vaccines (GP-Cda1, GP-Cda2, GP-Cda3, and GP-Sod1) afforded a significant survival advantage in at least one mouse model; some vaccine combinations provided added protection over that seen with either antigen alone. Vaccine-mediated protection against C. neoformans did not necessarily predict protection against C. gattii Vaccinated mice developed pulmonary inflammatory responses that effectively contained the infection; many surviving mice developed sterilizing immunity. Predicted T helper cell epitopes differed between mouse strains and in the degree to which they matched epitopes predicted in humans. Thus, we have discovered cryptococcal proteins that make promising candidate vaccine antigens. Protection varied depending on the mouse strain and cryptococcal species, suggesting that a successful human subunit vaccine will need to contain multiple antigens, including ones that are species specific.IMPORTANCE The encapsulated fungi Cryptococcus neoformans and Cryptococcus gattii are responsible for nearly 200,000 deaths annually, mostly in immunocompromised individuals. An effective vaccine could substantially reduce the burden of cryptococcosis. However, a major gap in cryptococcal vaccine development has been the discovery of protective antigens to use in vaccines. Here, six cryptococcal proteins with potential as vaccine antigens were expressed recombinantly and purified. Mice were then vaccinated with glucan particle preparations containing each antigen. Of the six candidate vaccines, four protected mice from a lethal cryptococcal challenge. However, the degree of protection varied as a function of mouse strain and cryptococcal species. These preclinical studies identify cryptococcal proteins that could serve as candidate vaccine antigens and provide a proof of principle regarding the feasibility of protein antigen-based vaccines to protect against cryptococcosis.
Keywords: Cryptococcus; bioinformatics; glucans; immunization; major histocompatibility complex.
Copyright © 2017 Specht et al.
Figures




References
-
- Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A (ed). 2011, Cryptococcus: from human pathogen to model yeast. ASM Press, Washington, DC.
-
- Byrnes EJ III, Li W, Lewit Y, Ma H, Voelz K, Ren P, Carter DA, Chaturvedi V, Bildfell RJ, May RC, Heitman J. 2010. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog 6:e1000850. doi: 10.1371/journal.ppat.1000850. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
